Team:KULeuven/Project/Filter
From 2008.igem.org
(Difference between revisions)
m |
|||
| Line 1: | Line 1: | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| - | <div style="float: right;">[[Image:pictogram_filter.png| | + | <div style="float: right;">[[Image:pictogram_filter.png|120px]]</div> |
==Filter== | ==Filter== | ||
| + | ===BioBricks=== | ||
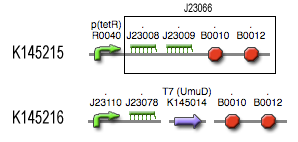
[[Image:Filter_BioBrick.jpg|center]] | [[Image:Filter_BioBrick.jpg|center]] | ||
| - | The filter | + | ===Components=== |
| + | [[Image:T7RNAP.png|290px|right|T7 RNA polymerase, N-terminal green, C-terminal yellow]]The filter, acting as a feedforward loop with AND-gate and therefor needs three specific components. | ||
| + | * The ''first'' is a fast degrading primary messenger. For this, we used [http://partsregistry.org/Part:BBa_J23009 '''BBa_J23009'''], the RiboKey3d (more information [http://partsregistry.org/Featured_Parts:RNA-Lock-and-Key here]). As this is an mRNA molecule, degradation occurs quite fast ('''referentie'''). | ||
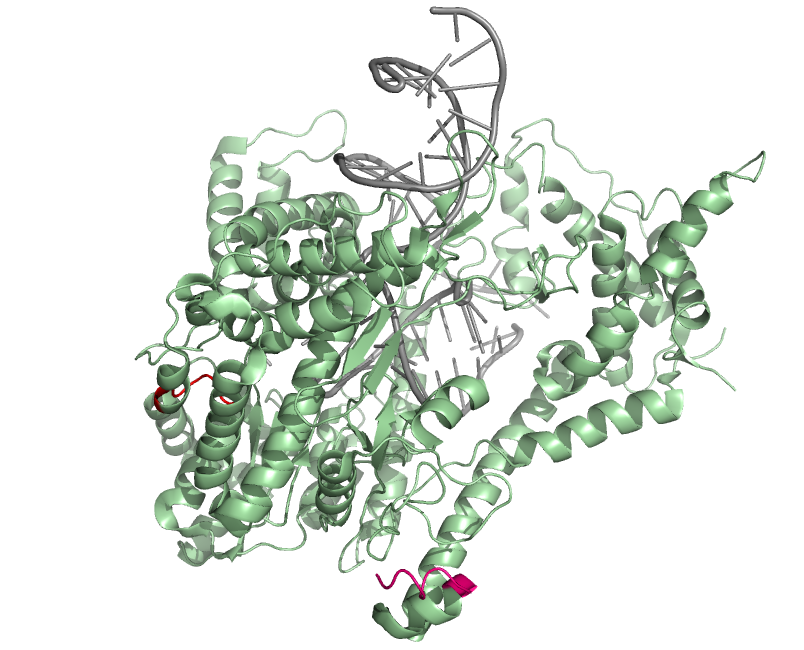
| + | * The ''second'' component of the system is a stable secondary messenger, that is induced by the first one. We opted for T7 RNA polymerase ([http://partsregistry.org/Part:BBa_K145xxx '''BBa_K145xxx''']). This molecule is much more stable than mRNA's, and modeling told us it was in fact too stable. So an UmuD derived N-terminal tag was added. Indeed, the crystal structure of T7 RNA polymerase showed catalytic activity at the C-terminus, which seems to be buried at the inside of the protein. The N-terminal is not catalytically active and also much more accessible. For more information on the UmuD-derived N-terminal tag, see the [https://2008.igem.org/Team:KULeuven/Literature literature]. | ||
| + | * The ''third'' and last specific component is an AND-gate. To make such a gate, we placed de filter output signal under control of the T7 promoter ([http://partsregistry.org/Part:BBa_I712074 '''BBa_I712074''']) and locked by the RiboLock3d ([http://partsregistry.org/Part:BBa_J23032 '''BBa_J23032''']). This AND-gate as such is not a part of the filter mechanism, but serves as promoter and RBS of parts that are dependent on the filter. | ||
| - | + | ===Action=== | |
| + | [[Image:filter_1.jpg|140px|left]]The components mentioned above now work in concert in what is called a coherent feedforward loop with AND-gate ([http://www.nature.com/ng/journal/v31/n1/abs/ng881.html reference]). Its action is particulary dependent on the above-mentioned differences in degradation time of the primary and secondary messenger. If background input signal is present, a small amount of primary messenger will result in a short amount of secondary messenger, but as a result of the time delay between the two, the primary signal will be degraded once the secondary signal is present. Because of this, the AND-gate will give no signal. Given a vast amount of input signal, the primary messenger will still be present once the secondary messenger is formed. This will lead to a nonzero signal from the AND-gate. | ||
| - | + | Indeed, production of the RiboKey3d will give rise to T7 RNA polymerase production. The T7 promoter and RiboLock3d together now form the AND-gate, giving transcription only when both the T7 RNA polymerase and RiboLock3d are present together. | |
| - | + | ||
| - | + | ||
For more information on the exact kinetics and dynamics of this system, see the modeling page ('''link'''). | For more information on the exact kinetics and dynamics of this system, see the modeling page ('''link'''). | ||
Revision as of 09:35, 31 July 2008
dock/undock dropdown
Contents |
Filter
BioBricks
Components
The filter, acting as a feedforward loop with AND-gate and therefor needs three specific components.- The first is a fast degrading primary messenger. For this, we used [http://partsregistry.org/Part:BBa_J23009 BBa_J23009], the RiboKey3d (more information [http://partsregistry.org/Featured_Parts:RNA-Lock-and-Key here]). As this is an mRNA molecule, degradation occurs quite fast (referentie).
- The second component of the system is a stable secondary messenger, that is induced by the first one. We opted for T7 RNA polymerase ([http://partsregistry.org/Part:BBa_K145xxx BBa_K145xxx]). This molecule is much more stable than mRNA's, and modeling told us it was in fact too stable. So an UmuD derived N-terminal tag was added. Indeed, the crystal structure of T7 RNA polymerase showed catalytic activity at the C-terminus, which seems to be buried at the inside of the protein. The N-terminal is not catalytically active and also much more accessible. For more information on the UmuD-derived N-terminal tag, see the literature.
- The third and last specific component is an AND-gate. To make such a gate, we placed de filter output signal under control of the T7 promoter ([http://partsregistry.org/Part:BBa_I712074 BBa_I712074]) and locked by the RiboLock3d ([http://partsregistry.org/Part:BBa_J23032 BBa_J23032]). This AND-gate as such is not a part of the filter mechanism, but serves as promoter and RBS of parts that are dependent on the filter.
Action
The components mentioned above now work in concert in what is called a coherent feedforward loop with AND-gate ([http://www.nature.com/ng/journal/v31/n1/abs/ng881.html reference]). Its action is particulary dependent on the above-mentioned differences in degradation time of the primary and secondary messenger. If background input signal is present, a small amount of primary messenger will result in a short amount of secondary messenger, but as a result of the time delay between the two, the primary signal will be degraded once the secondary signal is present. Because of this, the AND-gate will give no signal. Given a vast amount of input signal, the primary messenger will still be present once the secondary messenger is formed. This will lead to a nonzero signal from the AND-gate.Indeed, production of the RiboKey3d will give rise to T7 RNA polymerase production. The T7 promoter and RiboLock3d together now form the AND-gate, giving transcription only when both the T7 RNA polymerase and RiboLock3d are present together.
For more information on the exact kinetics and dynamics of this system, see the modeling page (link).
 "
"