Team:Paris/Modeling/Synchronisation
From 2008.igem.org
(→The Circuit) |
(→The Circuit) |
||
| Line 9: | Line 9: | ||
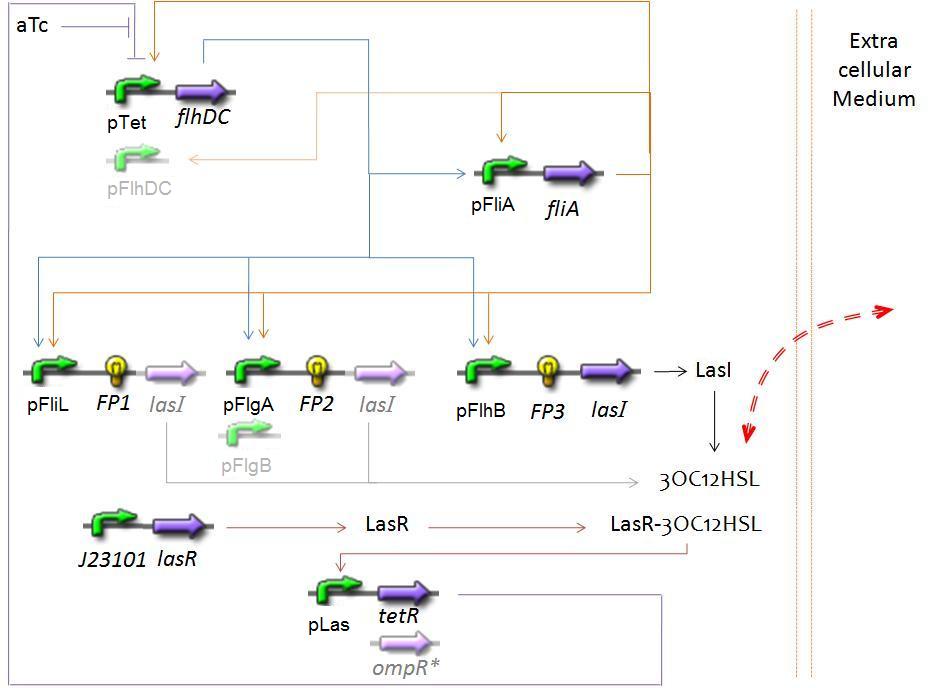
We will study three alternatives for the place of the ''lasI'' operon : on the first, second or third activated operon of the FIFO order. | We will study three alternatives for the place of the ''lasI'' operon : on the first, second or third activated operon of the FIFO order. | ||
| - | <center>[[Image: | + | <center>[[Image:Model_Synchrob.jpg|center]]</center> |
==Biochemical Assumptions== | ==Biochemical Assumptions== | ||
Revision as of 14:06, 19 August 2008
SynchronisationThe CircuitWe are know studying the following circuit, based on the one constituing the FIFO system, but with the mediation of AHL in the negative feed back loop. We will study three alternatives for the place of the lasI operon : on the first, second or third activated operon of the FIFO order. Biochemical Assumptionssee FIFO system As our medium is under a constant shaking, in an aqueous medium, we simulate the diffusion phenomenon as following : we suppose that the extra cellular medium has an homogeneous concentration of AHL, evolving along time. Then, for each cell, we will simply consider exchanges with this extre cellular medium, proportional to gradient. We will most probably use parameters found in literature for the diffusion constant. Resulting EquationsFirst, we kept the equations of the FIFO system. We must ignore the equations specifics to the "direct" feed back loop, without using "quorum sensing". Then, we added at the end of the previous document the added equation (beginning page 13), regard to the complexations and diffusions. Deterministic vs. Stochastic ModelFirst, we will simulate the resulting behaviour for only one cell. If this deterministic model leads to interesting oscillations, we will then simulate what happen for 10, 103, 106 (!), cells, to see if synchronisation is really effective. As the previous model is deterministic, we have to introduce differences between our cells. In this aim, we will use the standard deviation of our own measurements (see estimation of parameters) to give different parameters for each cell we introduce in our virtual medium... |
 "
"