Team:KULeuven/Model/Filter
From 2008.igem.org
(Difference between revisions)
(→Position in the system) |
(→Matlab (SBML file)) |
||
| Line 95: | Line 95: | ||
==== Matlab ([https://static.igem.org/mediawiki/2008/c/ce/Filter_Matlab.zip SBML file])==== | ==== Matlab ([https://static.igem.org/mediawiki/2008/c/ce/Filter_Matlab.zip SBML file])==== | ||
| - | |||
| - | |||
[[Image:Filter_Matlab.jpg|center]] | [[Image:Filter_Matlab.jpg|center]] | ||
| - | |||
=== Simulations === | === Simulations === | ||
Revision as of 09:19, 17 August 2008
dock/undock dropdown
Contents |
Filter
Position in the system
todo
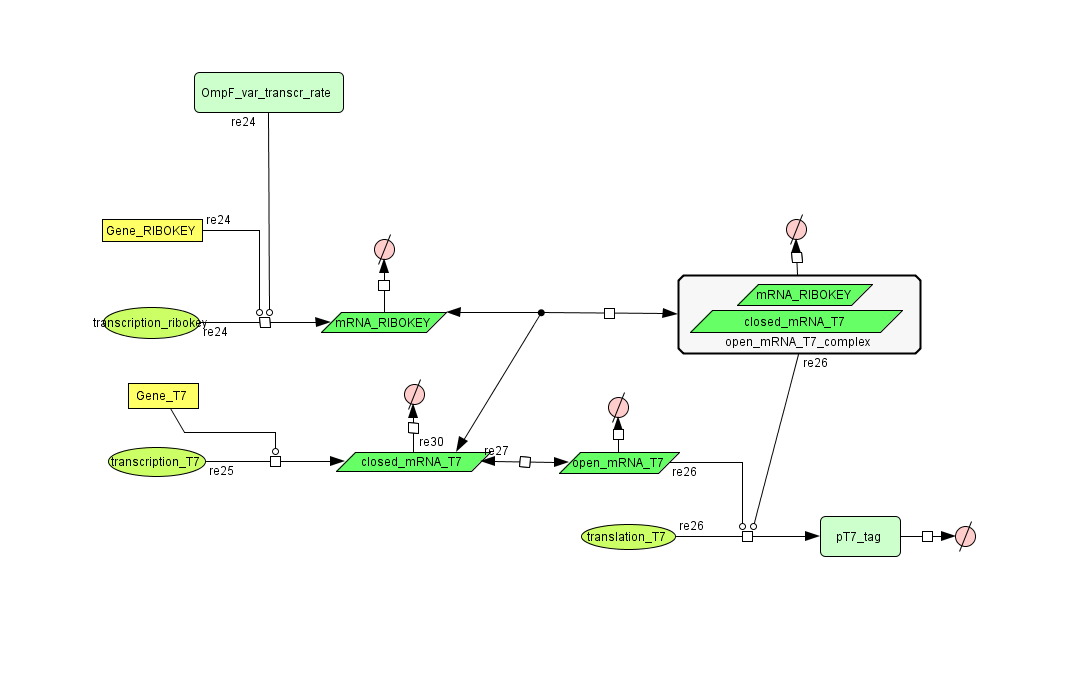
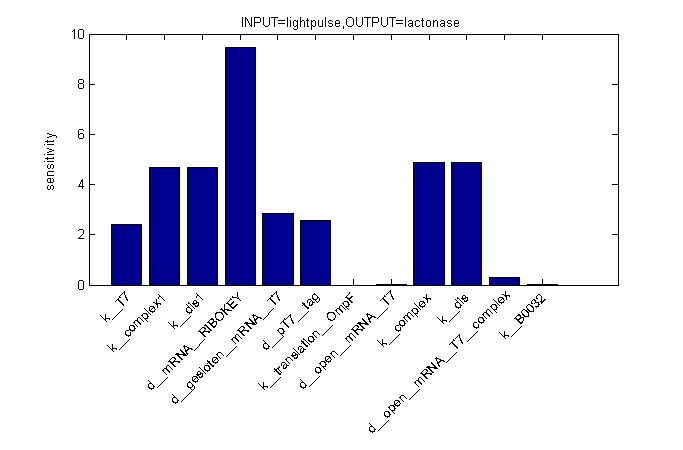
Describing the system
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation rates | |||
| dpT7_tag | 0.00155 s-1 | tag added to speed up degradation of otherwise too stable T7 polymerase | [http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP#Model_and_simulation link] |
| dmRNA_RIBOKEY | 0.00462 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dclosed_mRNA_T7 | 0.00462 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen_mRNA_T7 | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen_mRNA_T7_complex | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| Equilibrium constants | |||
| Keq_1 | 0,015 [M] | closed and open mRNA | |
| Keq_2 | 0.0212 [M] | closed and key (binded with ribokey) complex mRNA | |
| Transcription rates | |||
| kmRNA_RIBOKEY | Input dependent | ||
| kmRNA_T7 | 0,0011 s-1 | ||
Remark: The key-lock system can be enhanced to 0.3%-14% (todo: Jonas/Dries)
Models
CellDesigner (SBML file)
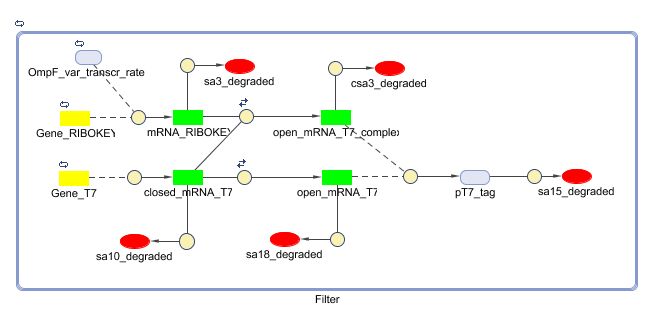
Matlab (SBML file)
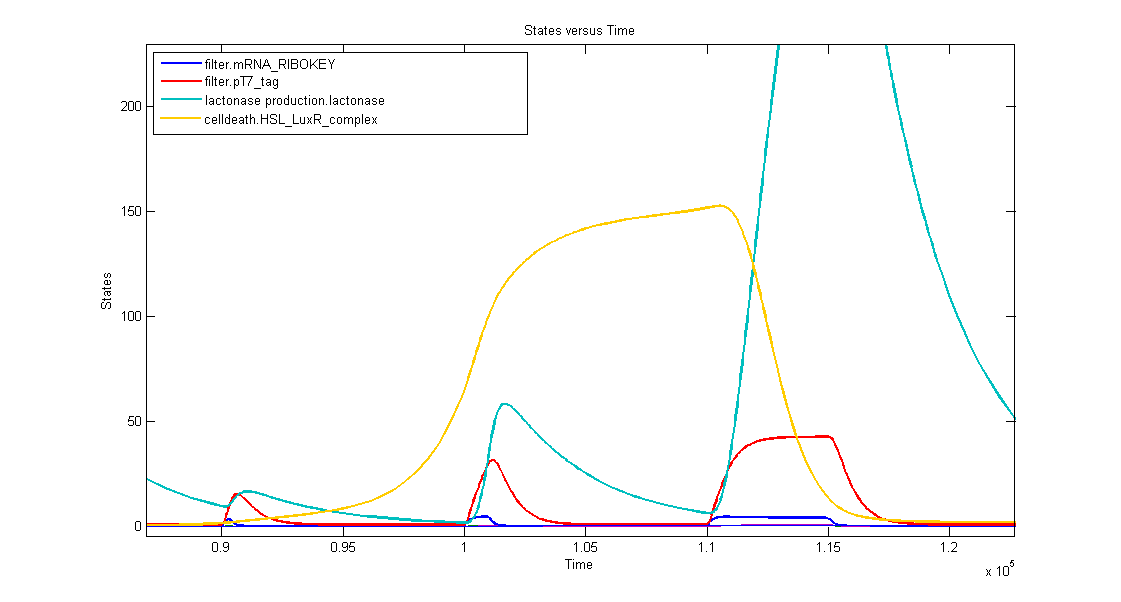
Simulations
Discussion of the simulation:
1. AND gate of the filter
In the simulation we can clearly see this series of events:
- when dark blue(ribokey) starts to increase, red (T7) also starts to increase, giving rise to an increasing amount of lactonase (blue) = AND-GATE.
- when dark blue(ribokey) starts to decrease, red (T7) also starts to decrease, but much slower. The lactonase also starts to decrease, as it should be.
The short lifetime of the ribokey compared to the lifetime of the T7-protein, guarantees that the AND-GATE always works perfectly fine: when there's no more input, the ribokey will rapidly decrease (and disappear) and makes sure that the AND-GATE is not activated anymore, even when the T7-protein is slowly decreasing.
2. Filtering in practice
- First Lightpulse: 300s
- small peak of lactonase
- no influence on the timer
- Second Lightpulse: 1000s
- medium peak of lactonase
- influences the timer by levelling the timing capabilities, but it doesn't reset the timer
- Third Lightpulse: 5000s
- huge peak of lactonase
- reset of the timer: amount of complex goes to zero
 "
"