Team:KULeuven/Model/Filter
From 2008.igem.org
(Difference between revisions)
(→Describing the system) |
(→Parameters) |
||
| Line 66: | Line 66: | ||
| K<sub>eq_1</sub> | | K<sub>eq_1</sub> | ||
| 0,015 [M] | | 0,015 [M] | ||
| - | | closed and open mRNA | + | | closed and open mRNA, derived from experiments |
| - | | | + | | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
|- | |- | ||
| K<sub>eq_2</sub> | | K<sub>eq_2</sub> | ||
| 0.0212 [M] | | 0.0212 [M] | ||
| - | | closed and key | + | | closed and key unlocked mRNA complex, derived from experiments |
| + | | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] | ||
| + | |- | ||
| + | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Rate constants | ||
| + | |- | ||
| + | | k<sub>dis</sub> | ||
| + | | 100 s<sup>-1</sup> | ||
| + | | derived from experimental values | ||
| + | | | ||
| + | |- | ||
| + | | k<sub>complex</sub> | ||
| + | | 57 s<sup>-1</sup> | ||
| + | | derived from experimental values | ||
| | | | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Transcription rates | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Transcription rates | ||
|- | |- | ||
| - | | | + | | TetR_var_transcr_rate |
| - | | | + | | p(TetR) dependent |
| - | | | + | | (RiboKey) between 5E-5 and 0.0125 s<sup>-1</sup> ~ [aTc] |
| - | | | + | | |
|- | |- | ||
| k<sub>mRNA_T7</sub> | | k<sub>mRNA_T7</sub> | ||
| Line 88: | Line 100: | ||
<b>Remark:</b> The key-lock system has been enhanced to 0.3%-14% (todo: new parameters will be added in a new overview) | <b>Remark:</b> The key-lock system has been enhanced to 0.3%-14% (todo: new parameters will be added in a new overview) | ||
| + | <!--DONE: k_dis, k_complex, TetR_var... , d_mRNA_ribokey, --!> | ||
=== Models === | === Models === | ||
Revision as of 10:35, 27 August 2008
dock/undock dropdown
Contents |
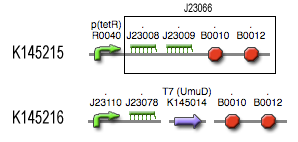
Filter
Position in the system
The filter is positioned immediately after the input, because its job is to filter out possible noise signals or background signals that aren't caused by the "desease". It is the starting piece of the whole system, situated before the invertimer- and the reset-subsystem.
Describing the system
see also: Project:Filter
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation rates | |||
| dpT7_tag | 0.00155 s-1 | tag added to speed up degradation of otherwise too stable T7 polymerase | [http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP#Model_and_simulation link] |
| dmRNA_RIBOKEY | 0.00462 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dclosed_mRNA_T7 | 0.00462 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen_mRNA_T7 | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen_mRNA_T7_complex | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| Equilibrium constants | |||
| Keq_1 | 0,015 [M] | closed and open mRNA, derived from experiments | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| Keq_2 | 0.0212 [M] | closed and key unlocked mRNA complex, derived from experiments | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| Rate constants | |||
| kdis | 100 s-1 | derived from experimental values | |
| kcomplex | 57 s-1 | derived from experimental values | |
| Transcription rates | |||
| TetR_var_transcr_rate | p(TetR) dependent | (RiboKey) between 5E-5 and 0.0125 s-1 ~ [aTc] | |
| kmRNA_T7 | 0,0011 s-1 | ||
Remark: The key-lock system has been enhanced to 0.3%-14% (todo: new parameters will be added in a new overview)
 "
"