|
|
| Line 940: |
Line 940: |
| | <br> | | <br> |
| | Digestion Protocol <br> | | Digestion Protocol <br> |
| - | - about 2µg Plasmid-Prep in 20µl <br>
| + | - about 2µg Plasmid-Prep in 20µl <br> |
| - | - 2 µl NEB Buffer 10x<br>
| + | - 2 µl NEB Buffer 10x<br> |
| - | - 1 µl NEB enzyme 1 (NgoMIV, AgeI, XbaI, EcoRI)<br>
| + | - 1 µl NEB enzyme 1 (NgoMIV, AgeI, XbaI, EcoRI)<br> |
| - | - 1 µl NEB enzyme 2 (PstI, SpeI)<br>
| + | - 1 µl NEB enzyme 2 (PstI, SpeI)<br> |
| - | - 0,2µl BSA 100x<br>
| + | - 0,2µl BSA 100x<br> |
| | <br> | | <br> |
| | Ligation<br> | | Ligation<br> |
| - | - 10µl volume of vector and insert DNA (about 50ng vector-DNA)<br>
| + | - 10µl volume of vector and insert DNA (about 50ng vector-DNA)<br> |
| - | - 1 µl DNA Quick Ligase (New England Biolabs)<br>
| + | - 1 µl DNA Quick Ligase (New England Biolabs)<br> |
| - | - 10 µl Quick Ligase Buffer<br>
| + | - 10 µl Quick Ligase Buffer<br> |
| | <br> | | <br> |
| | Analytic digestion<br> | | Analytic digestion<br> |
| - | - about 0,5 µg Plasmid-DNA in 5µl<br>
| + | - about 0,5 µg Plasmid-DNA in 5µl<br> |
| - | - 5µl H2O<br>
| + | - 5µl H2O<br> |
| - | - 0,5 µl NotI<br>
| + | - 0,5 µl NotI<br> |
| - | - 1µl NEB-Buffer<br>
| + | - 1µl NEB-Buffer<br> |
| - | - 0,1 µl BSA<br>
| + | - 0,1 µl BSA<br> |
| | <br> | | <br> |
| | Transformation<br> | | Transformation<br> |
| - | Competent cells (100µl) werde defrosted on ice<br>
| + | - Competent cells (100µl) werde defrosted on ice<br> |
| - | 10µl of the ligation was added<br>
| + | - 10µl of the ligation was added<br> |
| - | DNA and cells werde mixed softly <br>
| + | - DNA and cells werde mixed softly <br> |
| - | Incubation on ice for 20-30 min<br>
| + | - Incubation on ice for 20-30 min<br> |
| - | Heat shock at 42°C for 40 sek <br>
| + | - Heat shock at 42°C for 40 sek <br> |
| - | cells were cooled down on ice for 5-10 min<br>
| + | - cells were cooled down on ice for 5-10 min<br> |
| - | 900µl sterile 2YT Medium was added<br>
| + | - 900µl sterile 2YT Medium was added<br> |
| - | Incubation at 37°C for 60-70 min (shaker)<br>
| + | - Incubation at 37°C for 60-70 min (shaker)<br> |
| - | cells werde plated on 2YT-agar-plates with antibiotics<br>
| + | - cells werde plated on 2YT-agar-plates with antibiotics<br> |
| | <br> | | <br> |
| | Klenow fill in reaction | | Klenow fill in reaction |
| - | - 25pmol forward primer
| + | - 25pmol forward primer |
| - | - 25pmol reverse primer
| + | - 25pmol reverse primer |
| - | - 0,5 µl Klenow-fragment without exonuclease activity (Fermentas)
| + | - 0,5 µl Klenow-fragment without exonuclease activity (Fermentas) |
| - | - 2µl Klenow Buffer
| + | - 2µl Klenow Buffer |
| - | - 1µl dNTPs
| + | - 1µl dNTPs |
| - | - Add H¬¬2O to a volume of 20µl
| + | - Add H¬¬2O to a volume of 20µl |
| - | program: 94°C for 3min, cool down to 37°C, adition of klenow enzyme, 37°C for 1 hour
| + | program: 94°C for 3min, cool down to 37°C, adition of klenow enzyme, 37°C for 1 hour |
| | | | |
| | | | |
|
Home
The Team
Project Report
Parts
Modeling
Notebook
Safety
CoLABoration
|
_cloning strategy
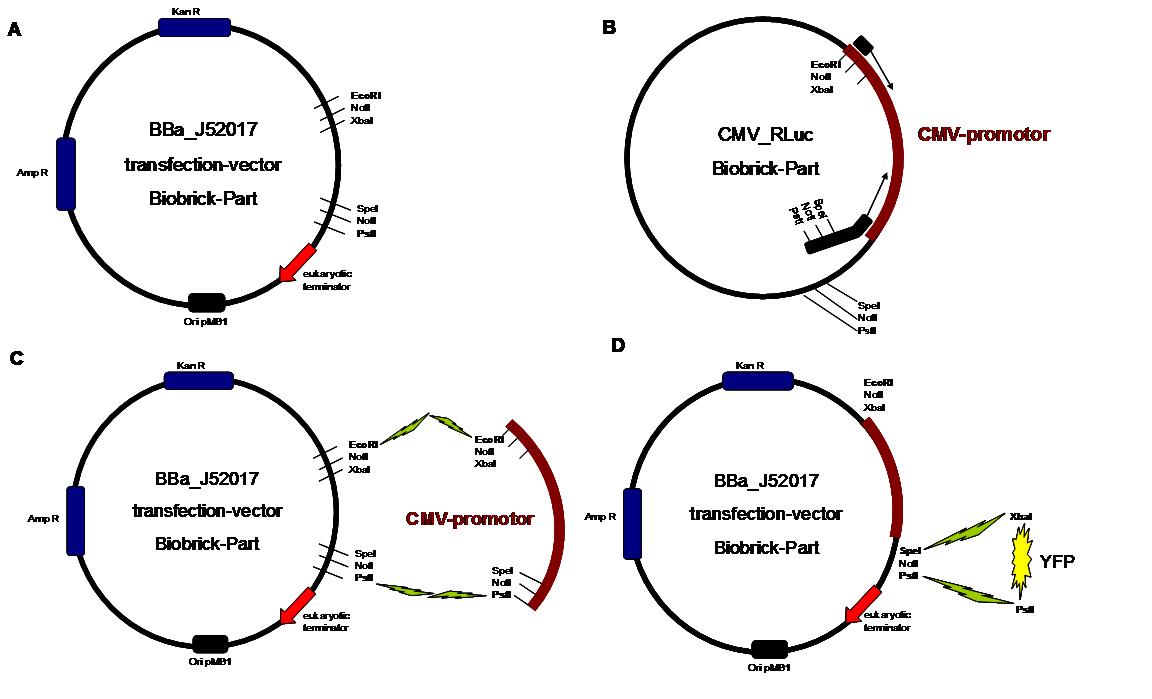
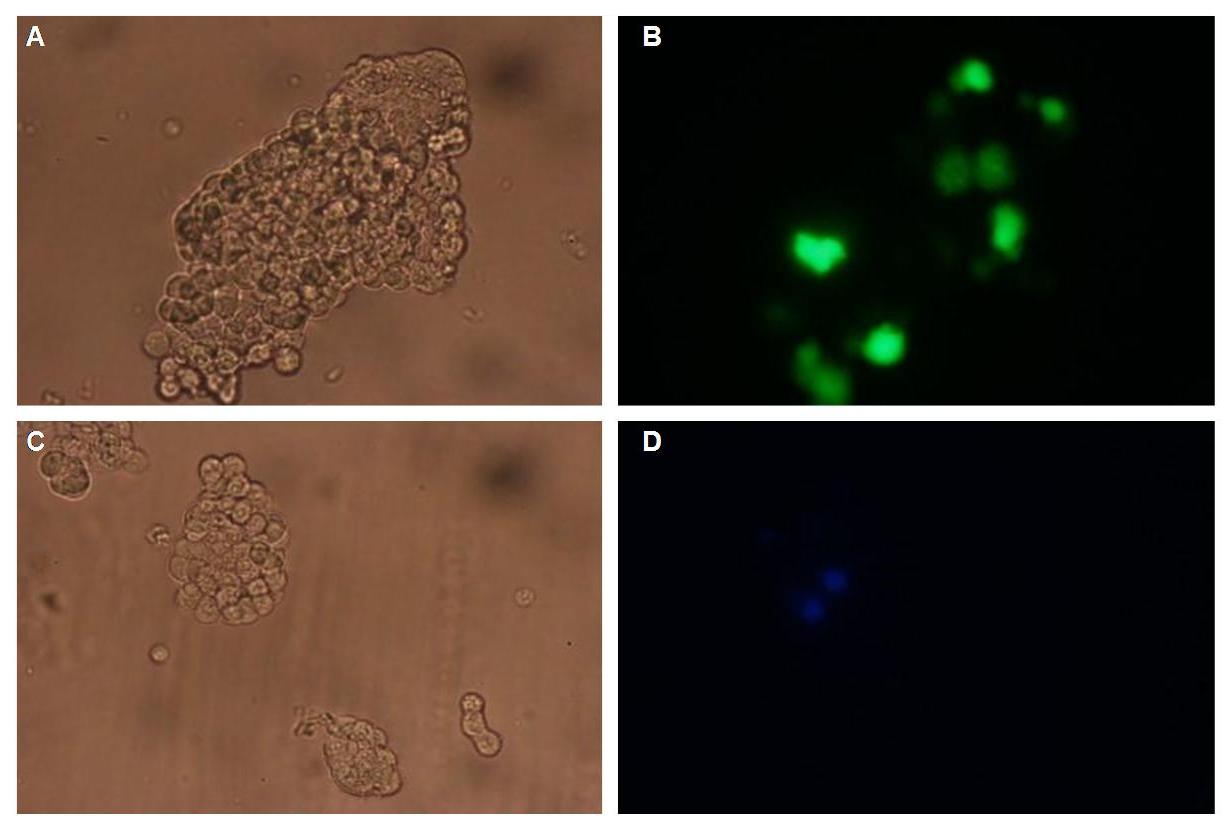
Figure 1 . A shows the Biobrick part BBa_J52017. The transfectionvektor for eukaryotic cell systems has an ampicillin- and a kanamycin resistance cassette. The multiple cloning site contains the Biobrick standard restriction sites EcoRI, NotI, XbaI, SpeI, NotI, PstI followed by an eukaryotic terminator sequence. B The CMV-promotor fragment was obtained by PCR with the Biobrick BBa-J52038 template. C The PCR product was cloned into the transfectionvector by EcoRI and PstI to get a final eukaryotic transfection-system. D To test the efficiency of expression a gene-fragment coding for the yellow fluorescent protein was put into the vector behind CMV-Promotor.
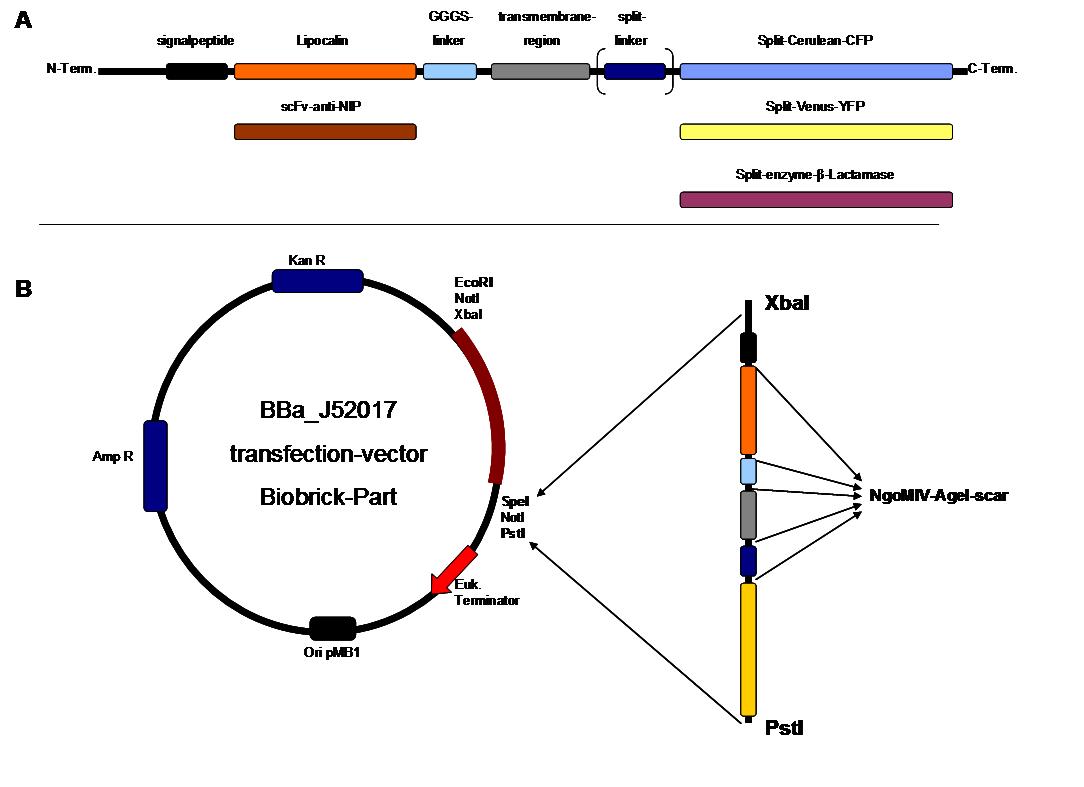
Figure 2 . A figure 2 A gives an overview about the cloning constructs. The N-terminal signal-peptide ensures protein transport to the cytoplasmamembrane. Lipocalin and the scFv-anti-NIP are the extracytoplasmatic parts of the construct to mediate signaltransduction into the cell. The GGGS-Linker keeps a distance to the transmembraneregion to overcome surface structures of the cell and to avoid a total inflexibility. Split fluorophor linker is only necessary for the C-terminal Split parts of Cerulean-CFP and Split-Venus-YFP. The Split enzymes β-Lactamase and Luciferase and the split-fluorophors CFP and YFP are the cytoplasmatic parts of the constructs. If there is a clustering of this synthetic receptor-system caused by the corresponding binding parts of Lipocalin and scFv-anti-NIP the split parts come together to create a functional protein, which allows a detection.
B The different constructions described in figure 2.A were cloned into the transfectionvector system by using the restriction sites XbaI and PstI to ensure a functional ATG-start codon which is part of the XbaI recognition-sequence in the iGEM-prefix
|
Step 1
|
Vector
digestion: EcoRI + PstI
|
Insert
digestion: EcoRI + PstI
|
|
|
BBa-J52017
|
_CMV-promotor
|
|
Step
2
|
Vector
digestion: AgeI+SpeI
|
Insert
digestion: NgoMIV+SpeI
|
|
|
pMA-BBFR
_ SPLIT-Linker
|
C-YFP
|
|
|
C-CFP
|
|
Step
3
|
Vector
digestion: AgeI+SpeI
|
Insert
digestion: NgoMIV+SpeI
|
|
|
pMA-BBFR
_egfR-Tm
|
_ N-β-Lactamase
|
|
|
_ C-β-Lactamase
|
|
|
_ SPLIT-Linker_ C-YFP
|
|
|
_ N-YFP
|
|
|
_ SPLIT-Linker_ C-CFP
|
|
|
_ N-CFP
|
|
|
_ BB058 (Luciferase)
|
|
|
_ BB057 (Luciferase)
|
|
Step
4
|
Vector
digestion: AgeI+SpeI
|
Insert
digestion: NgoMIV+SpeI
|
|
|
pMA-BBFR
_SP
|
_scFv-anti-NIP
|
|
|
_ Lipocalin
|
|
Step
5
|
Vector
digestion: AgeI+SpeI
|
Insert
digestion: NgoMIV+SpeI
|
|
|
pMA-BBFR
_SP_ scFv-anti-NIP
and
pMA-BBFR-+SP_ Lipocalin
|
_GGGS-linker (produced by Klenow fill in)
|
|
Step
6
|
Vector
digestion: AgeI+SpeI
|
Insert
digestion: NgoMIV+SpeI
|
|
|
pMA-BBFR
_SP_ scFv-anti-NIP _ GGGS-Li
and
pMA-BBFR
_ SP_ Lipocalin _
GGGS-Li
|
_
egfR-Tm _ N-β-Lactamase
|
|
|
_
egfR-Tm _ C-β-Lactamase
|
|
|
_
egfR-Tm _ SPLIT-Linker_ C-YFP
|
|
|
_
egfR-Tm _ N-YFP
|
|
|
_
egfR-Tm _ SPLIT-Linker_ C-CFP
|
|
|
_
egfR-Tm _ N-CFP
|
|
|
_
egfR-Tm _ BB058 (Luciferase)
|
|
|
_
egfR-Tm _ BB057 (Luciferase)
|
|
Step
7
|
Vector
digestion:
SpeI + PstI
|
Insert
digestion: XbaI + PstI
|
|
|
BBa-J52017_CMV
|
_SP_ scFv-anti-NIP_GGGS-Li_egfR-Tm_N-β-Lactamase
|
|
|
_ SP_ scFv-anti-NI _GGGS-Li_ egfR-Tm_C-β-Lactamase
|
|
|
_ SP_ scFv-anti-NIP_GGGS-Li_
egfR-Tm_SPLIT-Linker_C-YFP
|
|
|
_ SP_ scFv-anti-NIP_GGGS-Li_ egfR-Tm_N-YFP
|
|
|
_ SP_ scFv-anti-NIP_GGGS-Li_
egfR-Tm_SPLIT-Linker_C-CFP
|
|
|
_ SP_ scFv-anti-NIP_GGGS-Li_ egfR-Tm_N-CFP
|
|
|
_ SP_ scFv-anti-NIP_GGGS-Li _ egfR-Tm_BB058 (Luciferase)
|
|
|
_ SP_ scFv-anti-NIP_GGGS-Li _ egfR-Tm_BB057 (Luciferase)
|
|
|
_ SP_ Lipocalin _GGGS-Li_ egfR-Tm_N-β-Lactamase
|
|
|
_ SP_ Lipocalin _GGGS-Li_ egfR-Tm_C-β-Lactamase
|
|
|
_ SP_ Lipocalin _GGGS-Li_
egfR-Tm_SPLIT-Linker_ C-YFP
|
|
|
_ SP_ Lipocalin _GGGS-Li_
egfR-Tm_N-YFP
|
|
|
_ SP_ Lipocalin _GGGS-Li_
egfR-Tm_SPLIT-Linker_ C-CFP
|
|
|
_ SP_ Lipocalin _GGGS-Li_
egfR-Tm_N-CFP
|
|
|
_ SP_ Lipocalin _GGGS-Li__ egfR-Tm _ BB058 (Luciferase)
|
|
|
_ SP_ Lipocalin _GGGS-Li__ egfR-Tm _ BB057 (Luciferase)
|
[[METHODS]]
The cloning was started with a preparative digestion of the DNA-Plasmids. To clone fusion parts the vector constructs were digested with AgeI and PstI to open the Biobrick suffix. The inserts were digested with NgoMIV and PstI. For cloning into the transfection-vector the enzymes SpeI and PstI were used for vector and XbaI, PstI for insert to keep up the ATG-start codon in the XbaI restriction site of the biobrick suffix. All restriction-enzymes were ordered from New England Biolabs. After digestion the DNA-fragments were separated on a 1% agarose gel. The DNA-band of interest was isolated and purified with the QIAGEN QIAquick Gel Extraction Kit. For the ligation a 3 molar excess of the insert was put together with the vector-fragment and ligated with a Quick ligase (New England Biolabs). After half an our at room temperature the DNA was transformed to chemical competent E.coli strain XL1 cells, plated on 2YT-agar-plates and incubated at 37°C over night. After picking clones and growing in 5ml LB-medium, the plasmid DNA was isolated by QIAGEN QIAprep Spin Miniprep Kit. A test digestion was prepared with about 0,5µg Plasmid DNA and NotI restriction enzyme to isolate the fusion-protein from the vector and to control if the expected bands were obtained. After a positive result the clones were sent to GATC-Biotech for sequencing.
The GGGS-Linker was produced by Klenow -fill-in-PCR. Two primers were designed align to each other at 60°C and filled to a complete dobble-strand by Klenow Polymerase fragment.
Digestion Protocol
- about 2µg Plasmid-Prep in 20µl
- 2 µl NEB Buffer 10x
- 1 µl NEB enzyme 1 (NgoMIV, AgeI, XbaI, EcoRI)
- 1 µl NEB enzyme 2 (PstI, SpeI)
- 0,2µl BSA 100x
Ligation
- 10µl volume of vector and insert DNA (about 50ng vector-DNA)
- 1 µl DNA Quick Ligase (New England Biolabs)
- 10 µl Quick Ligase Buffer
Analytic digestion
- about 0,5 µg Plasmid-DNA in 5µl
- 5µl H2O
- 0,5 µl NotI
- 1µl NEB-Buffer
- 0,1 µl BSA
Transformation
- Competent cells (100µl) werde defrosted on ice
- 10µl of the ligation was added
- DNA and cells werde mixed softly
- Incubation on ice for 20-30 min
- Heat shock at 42°C for 40 sek
- cells were cooled down on ice for 5-10 min
- 900µl sterile 2YT Medium was added
- Incubation at 37°C for 60-70 min (shaker)
- cells werde plated on 2YT-agar-plates with antibiotics
Klenow fill in reaction
- 25pmol forward primer
- 25pmol reverse primer
- 0,5 µl Klenow-fragment without exonuclease activity (Fermentas)
- 2µl Klenow Buffer
- 1µl dNTPs
- Add H¬¬2O to a volume of 20µl
program: 94°C for 3min, cool down to 37°C, adition of klenow enzyme, 37°C for 1 hour

 
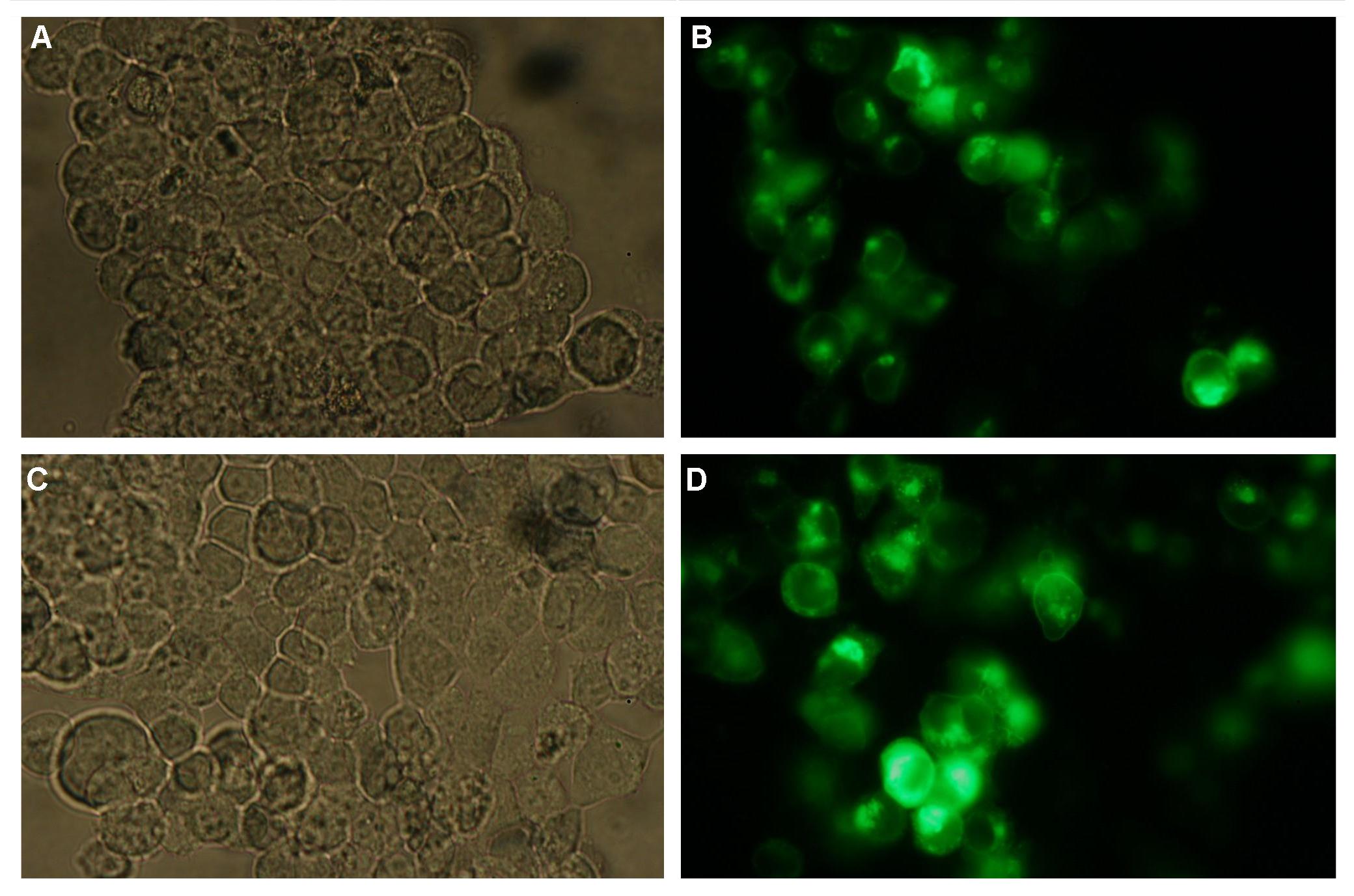
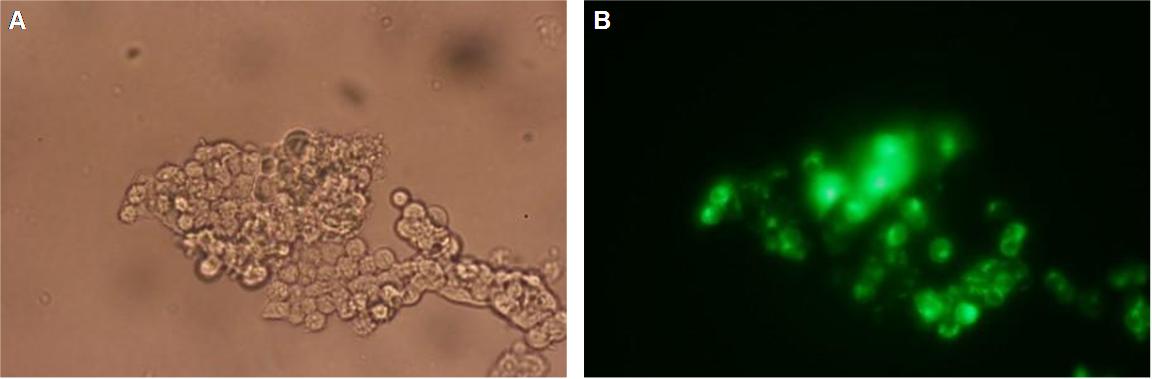
|
 "
"








