Team:UC Berkeley/LayeredAssembly
From 2008.igem.org
ChrisBrown (Talk | contribs) |
ChrisBrown (Talk | contribs) |
||
| Line 4: | Line 4: | ||
=='''Project Motivation'''== | =='''Project Motivation'''== | ||
| - | Imagine a world with no mini-preps. A world where DNA extraction and purification could be accomplished in a single eppendorf tube. We did, and so we created CloneBots, an automated approach to synthetic biology | + | Imagine a world with no mini-preps. A world where expensive cloning reagents were unnecessary. A world where DNA extraction and purification could be accomplished in a single eppendorf tube. We did, and so we created CloneBots, an automated approach to synthetic biology. |
| - | + | CloneBots seeks to simplify and automate the cloning process by creating protocols that involves only liquid handling steps that could more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the ''E. coli'', thereby allowing the cells to perform the required protocols in-vivo. | |
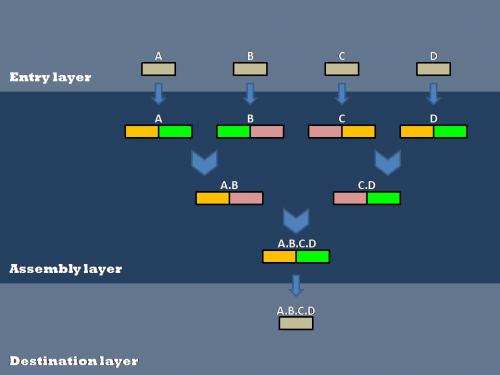
=='''Layered Assembly'''== | =='''Layered Assembly'''== | ||
| + | |||
| + | Our efforts to optimize synthetic biology protocols centered around the layered assembly scheme used by the Anderson Lab at UC Berkeley. Layered assembly is a straight-forward and robust method for combining two or more basic parts into a destination vector. There are three layers (Entry, Assembly and Destination). Both Entry and Assembly layers are standardized for ease of use, while the Destination plasmid can be tailored to a specific experiment or assay. | ||
[[Image:layered Assembly.jpg|center|frame|350px|A schematic representation of Layered Assembly]] | [[Image:layered Assembly.jpg|center|frame|350px|A schematic representation of Layered Assembly]] | ||
| - | |||
| - | |||
==='''Entry Layer'''=== | ==='''Entry Layer'''=== | ||
| - | The Entry | + | Biobrick parts are created in an entry vector. The Entry vector contains a Spectinomycin antibiotic resistance marker and EcoRI, BglII and BamHI restriction sites. The restriction sites are flanked by attR1 and attR2 recombination sites. |
| + | |||
[[Image: entry plasmdi]] | [[Image: entry plasmdi]] | ||
| - | Since the entry plasmid | + | Since the entry plasmid contains attR recombination sites, the basic part can easily be transferred into one of six different assembly vectors using the Gateway cloning scheme described here [[https://2008.igem.org/Team:UC_Berkeley/GatewayOverview]]. |
==='''Assembly Layer'''=== | ==='''Assembly Layer'''=== | ||
| - | The assembly layer is used to combine two or more basic parts in a specified order to create | + | The assembly layer is used to combine two or more basic parts in a specified order to create a composite part. |
| + | |||
| + | There are six assembly plasmids which contain two of three different antibiotic resistance markers (kanamycin, ampilicin and Chloramphenicol) separated by a XhoI restriction site. The basic parts are flanked by BglII and BamHI restriction sites. | ||
| + | |||
| + | The choice of antibiotic resistance markers in the assembly plasmids is predetermined such that once the two plasmids recombine, the new plasmid will have a combination of markers from the two assembly vectors. Although there are four possible products of this reaction, only one will have both of the correct antibiotic resistance markers. | ||
| + | |||
| + | The Clotho program developed by the UC Berkeley Comp team generates an assembly tree that minimizes the number of reactions required to make a composite part. To download Clotho, click here [[https://2008.igem.org/Team:UC_Berkeley_Tools/Project/Downloads]] | ||
| + | |||
| + | [[Image:Clothoscreenshot.jpg]] | ||
| + | |||
| + | The assembly plasmids are transformed into cells that are engineered to either methylate BamHI or BglII restriction sites. Part A is transformed into a "lefty" cell that is methylated on BglII restriction sites while Part B is transformed into a "righty" cell that is methylated on BamHI restriction sites. | ||
| + | |||
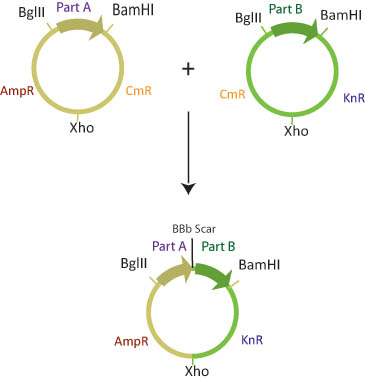
| + | In a single eppendorf tube, lefty and righty plasmid DNA is combined, along with BamHI, BglII, XhoI restriction enzymes and ligase. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The BamHI site in lefty and the BglII site in righty, and the Xho sites in both plasmids will be cut. Ligase will join the xho sites and the BglII/BamHI complimentary sticky ends to create a composite part with a different combination of antibiotic resistance genes than either parent plasmid. | ||
| + | |||
[[Image:cdb2ab.jpg]] | [[Image:cdb2ab.jpg]] | ||
| + | |||
| + | The assembly reaction can be repeated to combine a number of basic parts in a desired order. | ||
| + | |||
| + | Once the composite part is complete, it is transferred to a destination plasmid. | ||
==='''Destination Layer'''=== | ==='''Destination Layer'''=== | ||
| - | Completed composite parts are transferred from the assembly layer to the destination layer using the Gateway cloning scheme [[https://2008.igem.org/Team:UC_Berkeley/GatewayOverview]]. The | + | Completed composite parts are transferred from the assembly layer to the destination layer using the Gateway cloning scheme [[https://2008.igem.org/Team:UC_Berkeley/GatewayOverview]]. The destination plasmid is tailored to a specific experiment or assay. This allows a single composite part to be transfered to several different destination plasmids for further experimentation or characterization. |
[[Image:destplas]] | [[Image:destplas]] | ||
| Line 34: | Line 52: | ||
=='''Issues with Current Methods'''== | =='''Issues with Current Methods'''== | ||
| - | 1) Current protocols work well for BBa parts, but not well for BBb. CloneBots used BBb because BBa cloning requires several complex procedures while BBb has the potential to be fully automated. By using methylated plasmids, BBb assembly can be compartmentalized to a single tube or cell. | + | 1) Current protocols work well for BBa parts, but not as well for BBb. CloneBots used BBb because BBa cloning requires several complex procedures while BBb has the potential to be fully automated. By using methylated plasmids, BBb assembly can be compartmentalized to a single tube or cell. |
| - | 2) Gateway reactions, used for transferring basic parts into the assembly plasmids and transferring composite parts into destination plasmids, are expensive ($ | + | 2) Gateway reactions, used for transferring basic parts into the assembly plasmids and transferring composite parts into destination plasmids, are expensive ($8.65/reaction). |
3) Current protocols with no gel purification step are inefficient. | 3) Current protocols with no gel purification step are inefficient. | ||
| Line 46: | Line 64: | ||
5) People are more prone to make mistakes, i.e. switch tubes, use the wrong enzymes, etc. than robots. | 5) People are more prone to make mistakes, i.e. switch tubes, use the wrong enzymes, etc. than robots. | ||
| - | =='''CloneBots | + | =='''CloneBots Solutions'''== |
1) Engineer cells to express their own integrase (Int), excisionase (Xis), and integration host factor (IHF) enzymes to reduce the cost of Gateway reactions. For more information on in-vivo gateway, click here [[https://2008.igem.org/Team:UC_Berkeley/GatewayGenomic]]. | 1) Engineer cells to express their own integrase (Int), excisionase (Xis), and integration host factor (IHF) enzymes to reduce the cost of Gateway reactions. For more information on in-vivo gateway, click here [[https://2008.igem.org/Team:UC_Berkeley/GatewayGenomic]]. | ||
| - | 2) | + | 2) Eliminate mini-prep steps from cloning protocols by using our lysis device and in-vivo assembly. For more information on our lysis device, click here [[https://2008.igem.org/Team:UC_Berkeley/LysisDevice]]. |
| - | 3) | + | 3) Engineer cells and plasmids to express their own BamHI, BglII, Cre restriction enzymes and ligase so that assembly reactions in cell lysate and in-vivo are possible. For more information on assembly reactions in lysate or in-vivo, click here [[https://2008.igem.org/Team:UC_Berkeley/Assembly]]. |
| - | 4) By developing protocols that involve only liquid handling steps, cloning can be automated, requiring less labor. | + | 4) By developing protocols that involve only liquid handling steps, cloning can be automated, requiring less money and labor. |
Revision as of 00:34, 29 October 2008
Contents |
Project Motivation
Imagine a world with no mini-preps. A world where expensive cloning reagents were unnecessary. A world where DNA extraction and purification could be accomplished in a single eppendorf tube. We did, and so we created CloneBots, an automated approach to synthetic biology.
CloneBots seeks to simplify and automate the cloning process by creating protocols that involves only liquid handling steps that could more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the E. coli, thereby allowing the cells to perform the required protocols in-vivo.
Layered Assembly
Our efforts to optimize synthetic biology protocols centered around the layered assembly scheme used by the Anderson Lab at UC Berkeley. Layered assembly is a straight-forward and robust method for combining two or more basic parts into a destination vector. There are three layers (Entry, Assembly and Destination). Both Entry and Assembly layers are standardized for ease of use, while the Destination plasmid can be tailored to a specific experiment or assay.
Entry Layer
Biobrick parts are created in an entry vector. The Entry vector contains a Spectinomycin antibiotic resistance marker and EcoRI, BglII and BamHI restriction sites. The restriction sites are flanked by attR1 and attR2 recombination sites.
Since the entry plasmid contains attR recombination sites, the basic part can easily be transferred into one of six different assembly vectors using the Gateway cloning scheme described here [[1]].
Assembly Layer
The assembly layer is used to combine two or more basic parts in a specified order to create a composite part.
There are six assembly plasmids which contain two of three different antibiotic resistance markers (kanamycin, ampilicin and Chloramphenicol) separated by a XhoI restriction site. The basic parts are flanked by BglII and BamHI restriction sites.
The choice of antibiotic resistance markers in the assembly plasmids is predetermined such that once the two plasmids recombine, the new plasmid will have a combination of markers from the two assembly vectors. Although there are four possible products of this reaction, only one will have both of the correct antibiotic resistance markers.
The Clotho program developed by the UC Berkeley Comp team generates an assembly tree that minimizes the number of reactions required to make a composite part. To download Clotho, click here [[2]]
The assembly plasmids are transformed into cells that are engineered to either methylate BamHI or BglII restriction sites. Part A is transformed into a "lefty" cell that is methylated on BglII restriction sites while Part B is transformed into a "righty" cell that is methylated on BamHI restriction sites.
In a single eppendorf tube, lefty and righty plasmid DNA is combined, along with BamHI, BglII, XhoI restriction enzymes and ligase. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The BamHI site in lefty and the BglII site in righty, and the Xho sites in both plasmids will be cut. Ligase will join the xho sites and the BglII/BamHI complimentary sticky ends to create a composite part with a different combination of antibiotic resistance genes than either parent plasmid.
The assembly reaction can be repeated to combine a number of basic parts in a desired order.
Once the composite part is complete, it is transferred to a destination plasmid.
Destination Layer
Completed composite parts are transferred from the assembly layer to the destination layer using the Gateway cloning scheme [[3]]. The destination plasmid is tailored to a specific experiment or assay. This allows a single composite part to be transfered to several different destination plasmids for further experimentation or characterization.
Issues with Current Methods
1) Current protocols work well for BBa parts, but not as well for BBb. CloneBots used BBb because BBa cloning requires several complex procedures while BBb has the potential to be fully automated. By using methylated plasmids, BBb assembly can be compartmentalized to a single tube or cell.
2) Gateway reactions, used for transferring basic parts into the assembly plasmids and transferring composite parts into destination plasmids, are expensive ($8.65/reaction).
3) Current protocols with no gel purification step are inefficient.
Protocols involving gel purification and background subtraction steps to screen for the correct parts after assembly cannot be fully automated.
4) Mini-preps are time-consuming and expensive.
5) People are more prone to make mistakes, i.e. switch tubes, use the wrong enzymes, etc. than robots.
CloneBots Solutions
1) Engineer cells to express their own integrase (Int), excisionase (Xis), and integration host factor (IHF) enzymes to reduce the cost of Gateway reactions. For more information on in-vivo gateway, click here [[4]].
2) Eliminate mini-prep steps from cloning protocols by using our lysis device and in-vivo assembly. For more information on our lysis device, click here [[5]].
3) Engineer cells and plasmids to express their own BamHI, BglII, Cre restriction enzymes and ligase so that assembly reactions in cell lysate and in-vivo are possible. For more information on assembly reactions in lysate or in-vivo, click here [[6]].
4) By developing protocols that involve only liquid handling steps, cloning can be automated, requiring less money and labor.
 "
"