Team:KULeuven/Data/Other Components
From 2008.igem.org
(→Introduction) |
(→Construct & protocol) |
||
| Line 24: | Line 24: | ||
|} | |} | ||
<br> | <br> | ||
| - | Once this construct is made, testing the system becomes pretty straightforward. You would have to prepare multiple liquid cultures of the cells containing a plasmid with the above construct. | + | Once this construct is made, testing the system becomes pretty straightforward. You would have to prepare multiple liquid cultures of the cells containing a plasmid with the above construct. Then you can add aTc and remove it again after various time intervals (e.g. by transferring the cells to a fresh medium with no aTc). This will establish input signals of different lengths and you can detect the effect of each input signal by measuring the fluorescence of GFP (e.g. with FACS). You should detect no fluorescence when you gave a short input signal and you should be able to detect fluorescence when the input signal was longer. If you prepare various samples with various input lengths, you can make a plot of the fluorescence as a function of the input duration. This way you should be able to determine the threshold of the Filter (which signals does the Filter filter out, and which ones does he let through?). |
=== Results === | === Results === | ||
Revision as of 18:34, 29 October 2008
Contents |
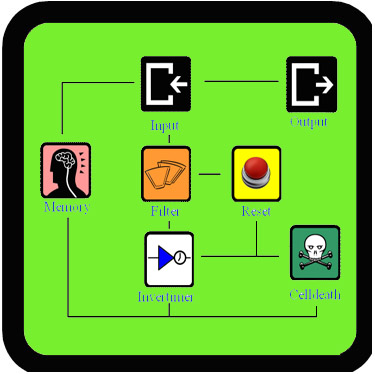
Introduction
As Dr.Coli is a very extensive project, there was not enough time to test all the new parts that we created. This page contains a summary of how our components can be tested (the constructs that have to be made and the actual testing protocol). It will also give an overview of the results we had so far. We worked as much parallel as possible, building different parts and subsystems at the same time. This way we could still have some results, even when some of the devices didn't succeed. Some of the constructs are already built, but we will leave the actual testing and finishing of the parts to the next year's teams, our advisors or you :-).
Filter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K145215 Parts Registry:K145215] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K145216 Parts Registry:K145216]
Construct & protocol
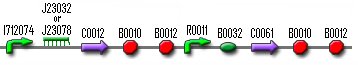
Our Filter is a composite part that is designed to filter out background noise of input. It can do this by a feedforward mechanism that uses an AND-gate. It works like this: if there is an input signal, the filter will first produce a ribokey. This key will then unlock a ribolock and this will lead to the expression of T7 RNA polymerase. Both this key and T7 RNA polymerase are the input of the AND-gate (which consists of a T7 promoter and ribolock) and both are necessary to get output from this AND-gate. So, when the input signal is very short, the ribokey will already be degraded by the time that T7 RNA polymerase is produced. In that case the AND-gate will not give an output. But when the input signal is longer, ribokey will still be available when T7 RNA polymerase is produced and therefore the AND-gate will produce an output. This way the filter can distinguish short signals (noise) from long signals (relevant signals).
So if we want to test the Filter, we need an input system (which is a constitutively produced TetR that will switch off the filter until aTc is added), the Filter and a GFP connected to the AND-gate:
| + | + | |||
| Input | Filter | AND-gate with GFP |
Once this construct is made, testing the system becomes pretty straightforward. You would have to prepare multiple liquid cultures of the cells containing a plasmid with the above construct. Then you can add aTc and remove it again after various time intervals (e.g. by transferring the cells to a fresh medium with no aTc). This will establish input signals of different lengths and you can detect the effect of each input signal by measuring the fluorescence of GFP (e.g. with FACS). You should detect no fluorescence when you gave a short input signal and you should be able to detect fluorescence when the input signal was longer. If you prepare various samples with various input lengths, you can make a plot of the fluorescence as a function of the input duration. This way you should be able to determine the threshold of the Filter (which signals does the Filter filter out, and which ones does he let through?).
Results
We did a lot of PCRs, digesting and ligating to prepare the construct. The input and the ANG-gate with GFP were properly constructed, as was the first half of the filter (R0040+J23066). But we had some problems with the second part of the filter. In the beginning the PCRs to prepare T7 polymerase failed over and over again. When it finally did succeed, we couldn't manage to insert this T7 RNA polymerase gene into the rest of the filter. So this test module was only partially built.
Memory
Construct & protocol
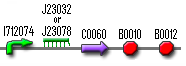
The Memory must keep the Cell Death inactivated until a decent input signal has been received; it is a stable switch that can only be switch on once (and then never go back). When there is no input signal, cII P22 will take control and it will repress cI 434, which is located under a cII P22 repressible promoter ([http://partsregistry.org/Part:BBa_R0053 R0053]). This is the OFF state in which the memory can remain indefinitely unless an input signal emerges. The first input signal will cause the production of cI 434 which will repress cII P22 production (because it is located under a cI 434 repressible promoter, [http://partsregistry.org/Part:BBa_R1052 R1052]), allowing cI 434 production to start from the cII P22 repressible promoter. After an input signal of about 1000 seconds, enough of the cII P22 has disappeared in order to make the cI 434 production self-sufficient. At this point, the memory has reached the ON state in which it will remain. So, schematically, the two possible states are:
- OFF state: cII P22 represses transcription from R0053 (no production cI 434)
- ON state: cI 434 represses transcription from R1052 (no production cII P22)
| + | + | |||
| Input | Memory | GFP-LVA controlled by cI 434 |
To test whether the Memory really works as a one-time stable switch, you should make liquid cultures of cells that contain the whole memory and a GFP-LVA controlled by cI 434 (see figure shown above). This means that GFP-LVA will only be produced when the memory is in the OFF state, thus in the beginning. When we give input (by adding aTc), the Memory switches to the ON state and the production of GFP-LVA stops. Because this GFP has an LVA tag it should be degraded very fast and you should be able to see the fluorescence decrease rapidly. To see if the ON state remains on, you can transfer the cells to a medium that doesn’t contain aTc. This means that there is no longer input, but the Memory should remain ON and so you should not detect any fluorescence. Each time the fluorescence can be measured with standard methods like e.g. FACS.
If you want an even more visual test procedure, you can add both a GFP-LVA controlled by cI 434 and a RFP controlled by cII P22. This means that, when the Memory is in the OFF state you should detect green fluorescence and when the Memory is in the ON state you should detect red fluorescence.
Results
The design of the Memory changed a lot during the three months we spent on this project. Therefore we didn’t have enough time to finish the latest update of the Memory. We decided not to construct it in the wet lab. However, it was very nicely modeled by the engineers, check it out here.
InverTimer, Reset, Cell Death
Construct & protocol
The InverTimer, Reset and Cell Death are essential subsystems that allow controlled cell death when Dr.Coli is no longer needed to cure the patient. This means that these parts are very tightly connected to each other and testing them separately would be very challenging. In order to test these systems, we would actually have to build the full model and then see how Dr.Coli reacts on different input signals (see full model). This could never be done within three months, but we started to build these parts anyway, so that someone else can finish it. These are the constructs:
| InverTimer: | |
| Reset: | |
| Cell Death: |
Results
In order to make the Cell Death construct, we had to make a hybrid promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K145150 K145150]) and the ccdB gene ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K145151 K145151]). This succeeded, but we couldn’t finish the whole construct.
The InverTimer and the Reset were completely built using the standard assembly method. But sequencing showed that we made a little mistake: we built in the wrong promoter. We then tried to undo this mistake by designing primers that can change the promoter to the correct one. This PCR succeeded, and so we digested the PCR products with EcoRI and XbaI and ligated it into pSB1A2. These plasmids were then electroporated into Top10, but we never got any colonies (we tried it a couple of times). We sent the other parts to the Registry anyway ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K145300 K245300] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K145302 K145302]); maybe someone else can use them in another project.
 "
"