Team:Freiburg/Project
From 2008.igem.org
m |
|||
| Line 8: | Line 8: | ||
'''Modular Synthetic Receptor System'''<br> | '''Modular Synthetic Receptor System'''<br> | ||
[[Image:Freiburg08 MSRS Schema_txt.png|thumb|right|250 px]] | [[Image:Freiburg08 MSRS Schema_txt.png|thumb|right|250 px]] | ||
| - | In the following we provide a summary of our project and display the highlights of our achievements. For a more detailed view please see the reports of each | + | In the following we provide a summary of our project and display the highlights of our achievements. For a more detailed view please see the reports of each subproject.<br><br> |
| + | |||
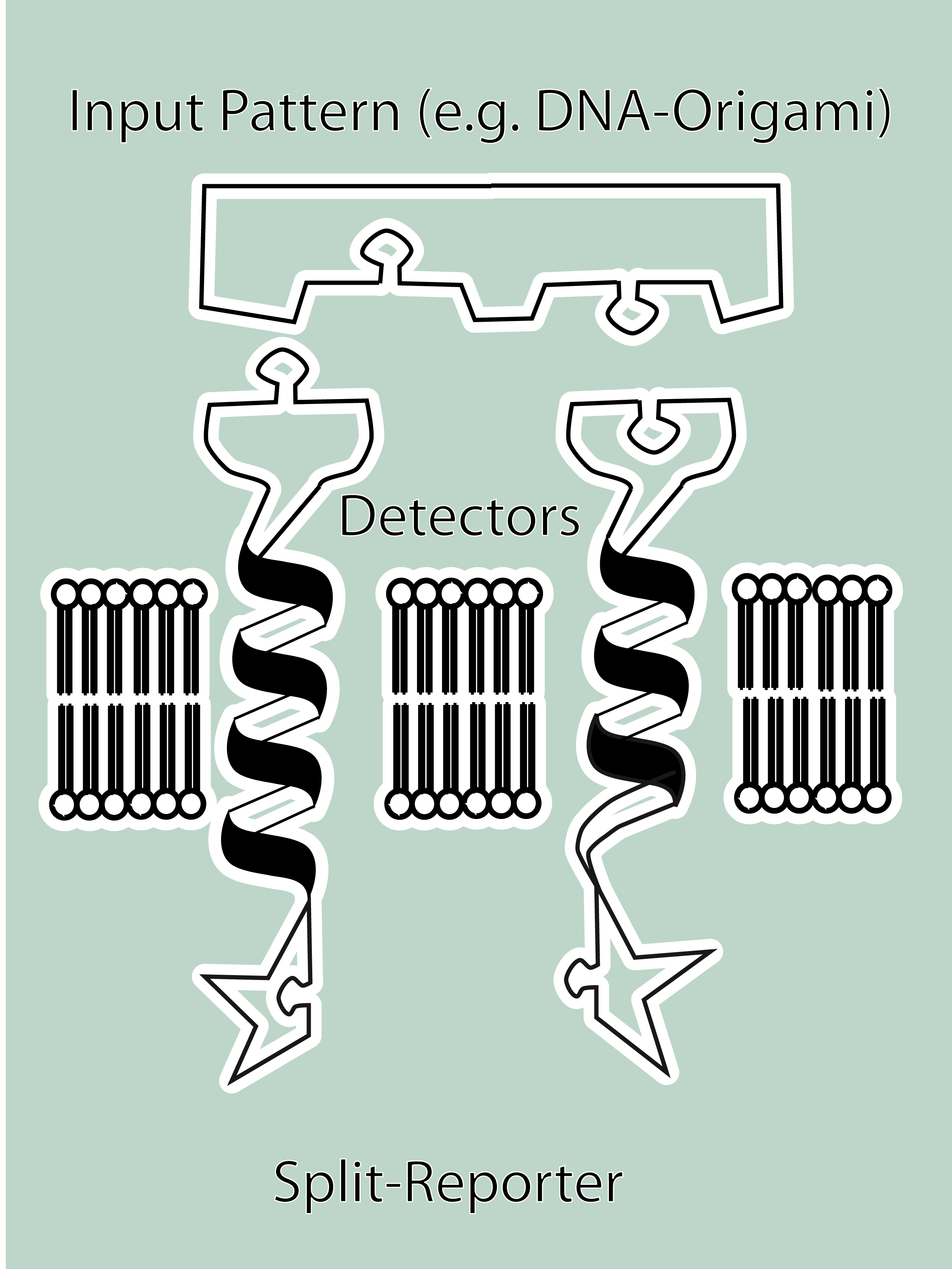
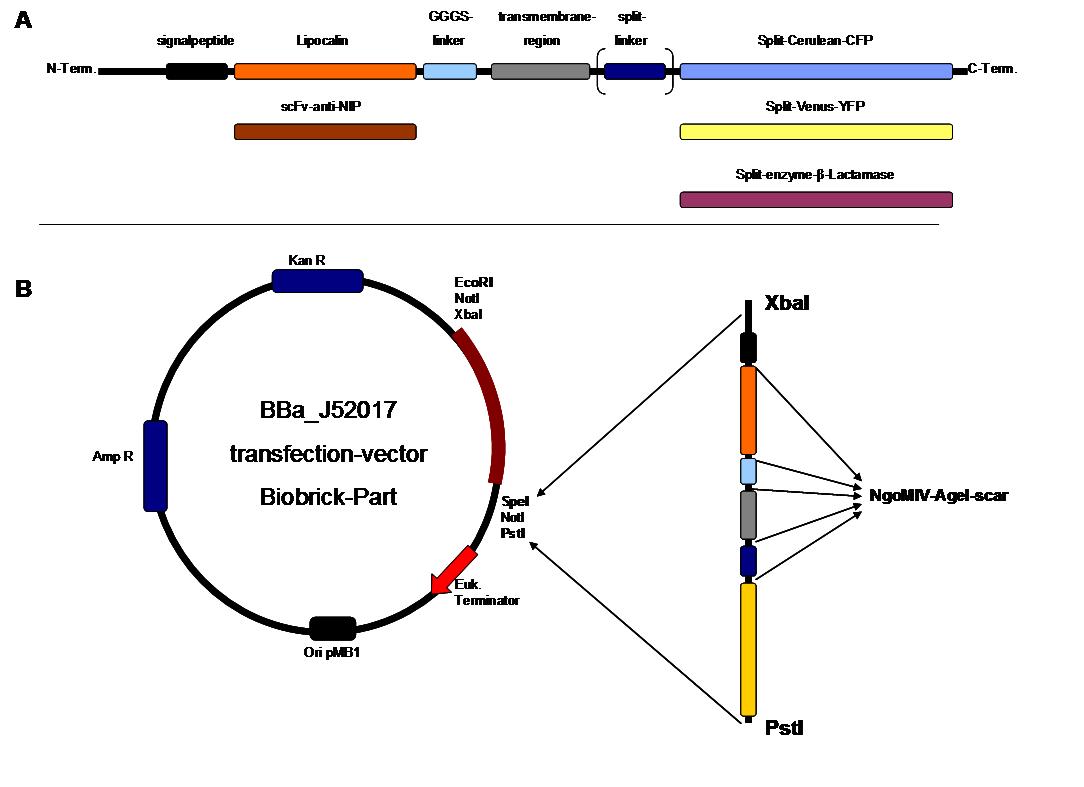
The general goal of the Freiburg 2008 team is to establish a synthetic transmembrane receptor system comprising the extracellular input devices, the extracellular receiver domain, the transducer to the cytosol and the reporter/executor in the cytosol.<br> | The general goal of the Freiburg 2008 team is to establish a synthetic transmembrane receptor system comprising the extracellular input devices, the extracellular receiver domain, the transducer to the cytosol and the reporter/executor in the cytosol.<br> | ||
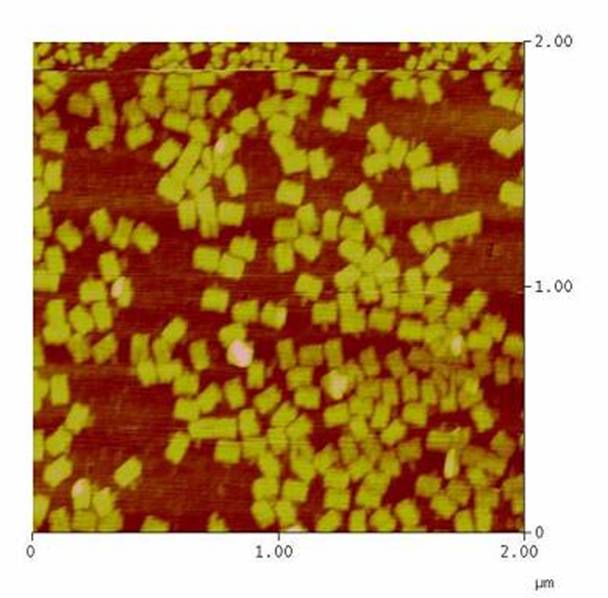
| - | As controllable receptor activation event, we chose spatial control of dimerization, which is a mechanism often used by nature. To achieve spatial control in the nanometer range | + | As controllable receptor activation event, we chose spatial control of dimerization, which is a mechanism often used by nature. To achieve spatial control in the nanometer range we chose DNA origami for exact and complex patterns with several ligands and we opted for chemical coupling of one type of ligand to a scaffold protein for simple multimerization.<br> |
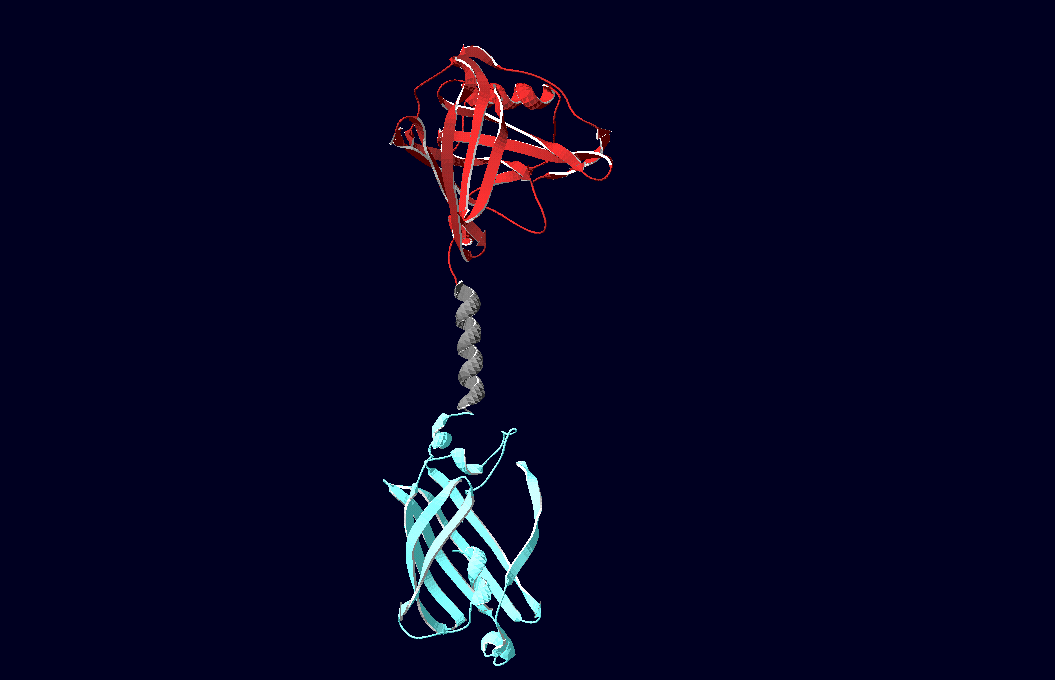
| - | To facilitate binding, we used a single-chain Fv fragment and a designed anticalin. Both bind haptens which can be conjugated to the DNA-Origami or scaffold proteins. Transduction to the cytosole is mediated by a single transmembrane helix.<br> | + | To facilitate binding, we used a single-chain Fv antibody fragment and a designed anticalin. Both bind haptens (nitro-jodo-phenol or fluorescein) which can be conjugated to specific oligonucleotides of the DNA-Origami or scaffold proteins. Transduction to the cytosole is mediated by a single transmembrane helix.<br> |
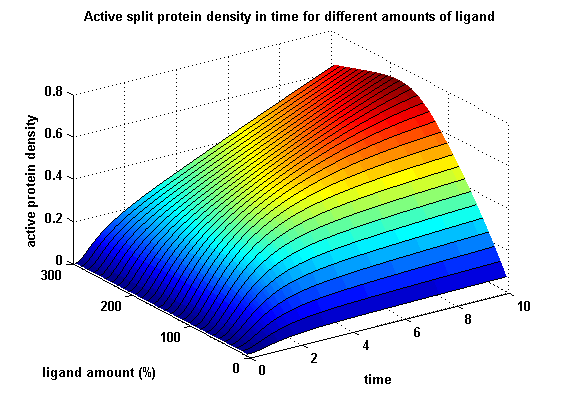
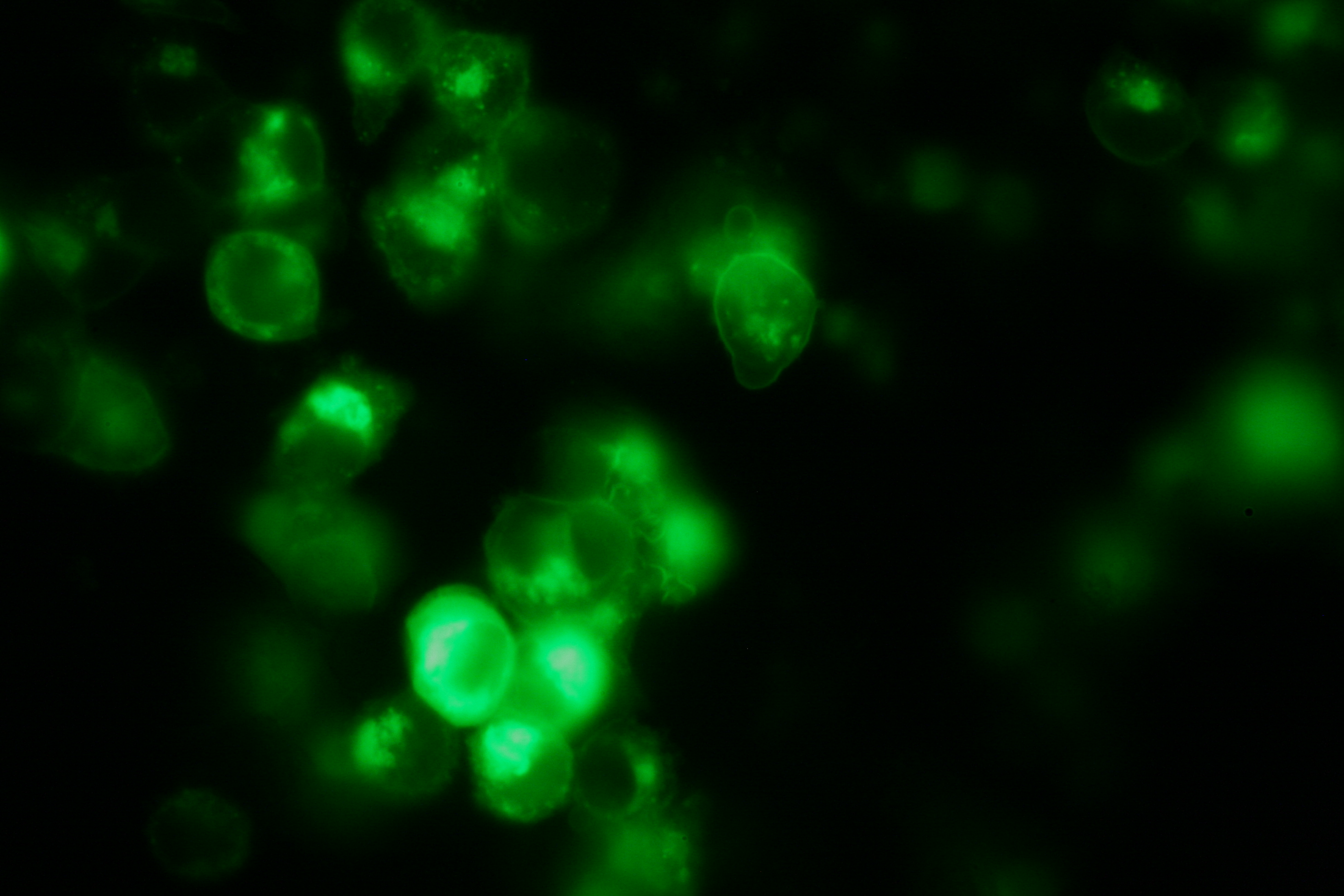
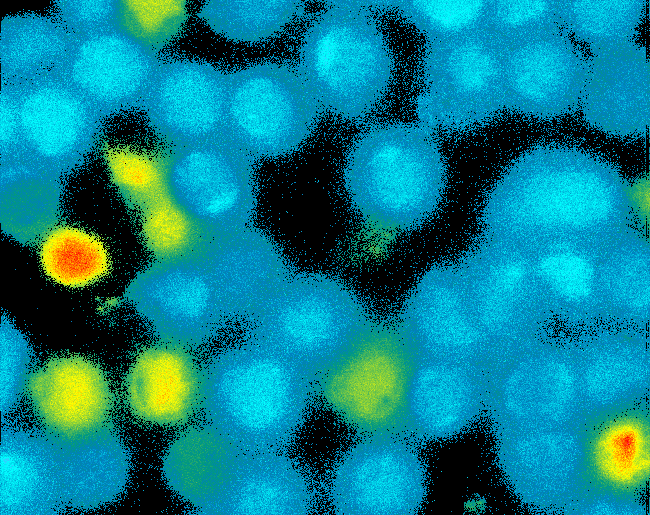
As intracellular reporters we employ split proteins. Here, we used the split lactamase from the iGEM2007 Freiburg project as well as the split fluorescent proteins CFP/cerulean and YFP/venus. To provide steric flexibility and reach of our binding and reporting modules we interspersed linkers between each functional domain.<br> | As intracellular reporters we employ split proteins. Here, we used the split lactamase from the iGEM2007 Freiburg project as well as the split fluorescent proteins CFP/cerulean and YFP/venus. To provide steric flexibility and reach of our binding and reporting modules we interspersed linkers between each functional domain.<br> | ||
| - | All parts | + | All parts feature full BioBrick-compatibility and in addition allow for the construction of fusion proteins due to extended Biobrick 3.0 standard. Cloning was done in <i>E. coli</i>. However, we test our receptors in human or mouse cells/chassis. Thus, to direct our constructs to the cell surface we attached a eukaryotic signal peptide to the N-terminus of each synthetic receptor. In addition to the new synthetic receptors, we also employed an existing T-cell line displaying a TCR fused to a single chain as a test for the spatial control.<br> |
Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiment. In particular we wanted to test the spatial control via DNA origami using an existing cell line expressing the scFv fused to a T-cell receptor. Here we addressed the following questions:<br><br> | Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiment. In particular we wanted to test the spatial control via DNA origami using an existing cell line expressing the scFv fused to a T-cell receptor. Here we addressed the following questions:<br><br> | ||
- Can we improve DNA origami assembly yield by varying the staple oligonucleotide to scaffold DNA ratio?<br> | - Can we improve DNA origami assembly yield by varying the staple oligonucleotide to scaffold DNA ratio?<br> | ||
Revision as of 23:20, 29 October 2008
|
Project Report |
_project report
SummaryModular Synthetic Receptor System In the following we provide a summary of our project and display the highlights of our achievements. For a more detailed view please see the reports of each subproject. The general goal of the Freiburg 2008 team is to establish a synthetic transmembrane receptor system comprising the extracellular input devices, the extracellular receiver domain, the transducer to the cytosol and the reporter/executor in the cytosol. SubprojectsModeling3D-ModelingDNA-OrigamiCloning StrategyTransfection and Synthetic Receptor ActivationCell Stability, Ca2+ Signaling and DNA-Origami-BindingHighlights
LiteratureSplit-fluorophores: |
 "
"