From 2008.igem.org
(Difference between revisions)
|
|
| Line 22: |
Line 22: |
| | </td> | | </td> |
| | <td> | | <td> |
| - | [[Image:Freiburg08_DNA-NIP5.png|thumb|left|380 px|'''Fig.3:''' Oligo-pattern with scale and two fragments (dimerization model)]] | + | [[Image:Freiburg08_DNA-NIP5.png|thumb|left|380 px|'''Fig.3:''' Oligo-pattern with scale and two fragments ]] |
| | </td> | | </td> |
| | </tr> | | </tr> |
Revision as of 01:30, 30 October 2008

|
Home
The Team
Project Report
Parts
Modeling
Notebook
Safety
CoLABoration
|
_3D-modeling
DNA-Origami
Planning the input pattern on the DNA-Origami surface, we generated various 3D-models of the huge molecule using "Nano-Engineer", Pymol and SwissPdbViewer (both freeware). Some of those models are shown here to give you an impression of the molecules we have engineered this year.
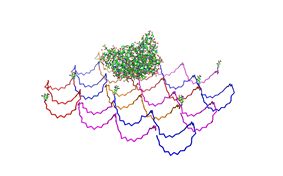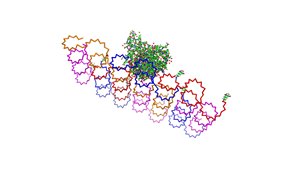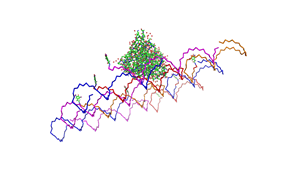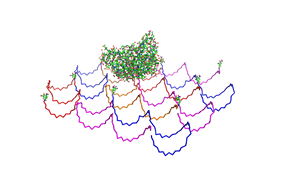 Fig.1: Some of the oligonucleotides that shape the single-stranded template fused to NIP-molecules and an anti-NIP-Fv-Fragment |
|
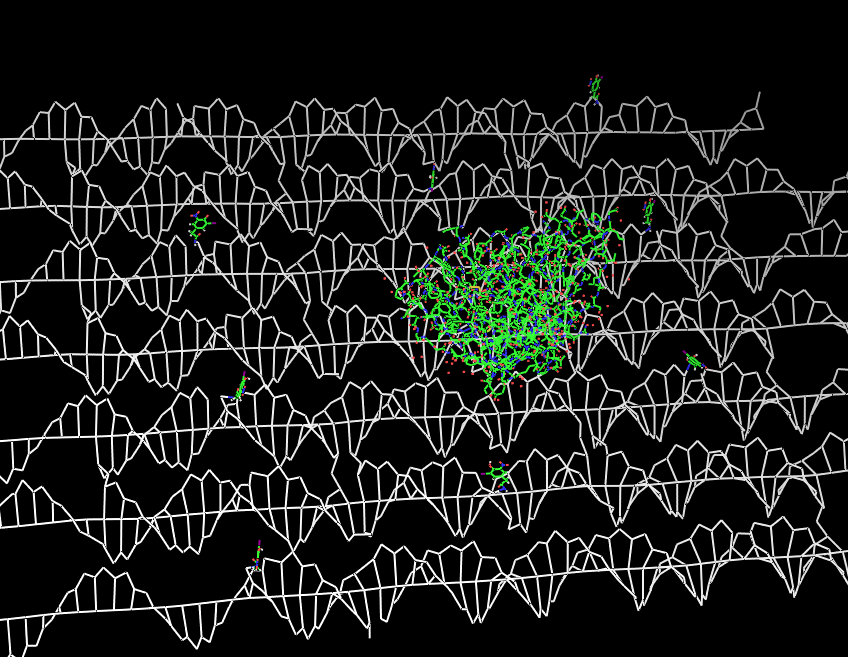 Fig.2: DNA-Origami with NIP-molecules and anti-NIP-fragment |
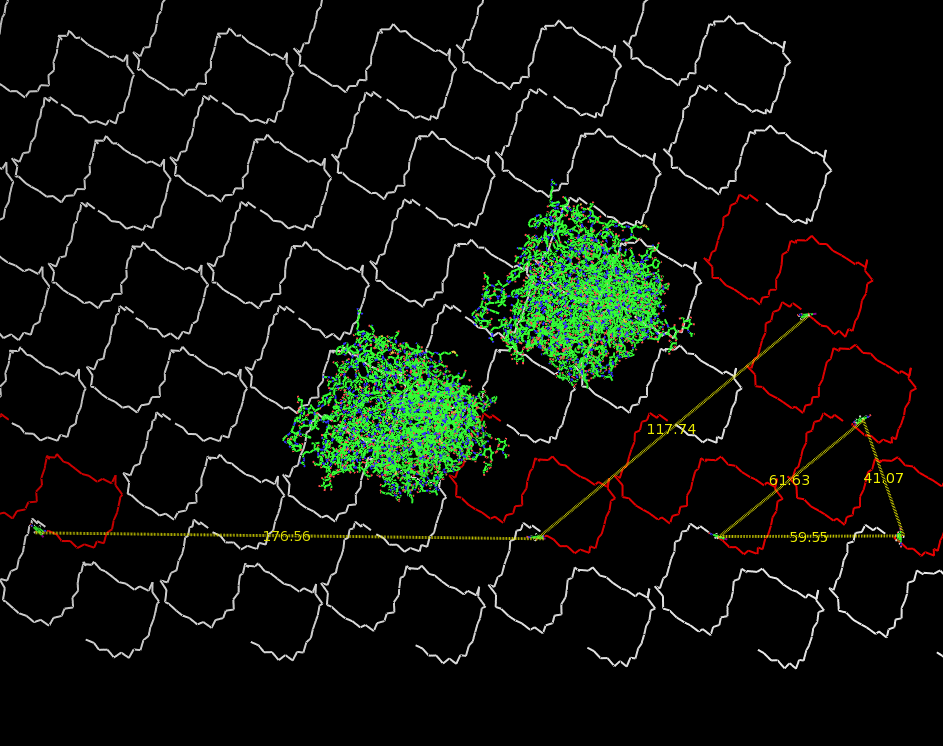 Fig.3: Oligo-pattern with scale and two fragments |
 Fig.4: Oligo-pattern with scale and two T-Cell-receptors (dimerization model) |
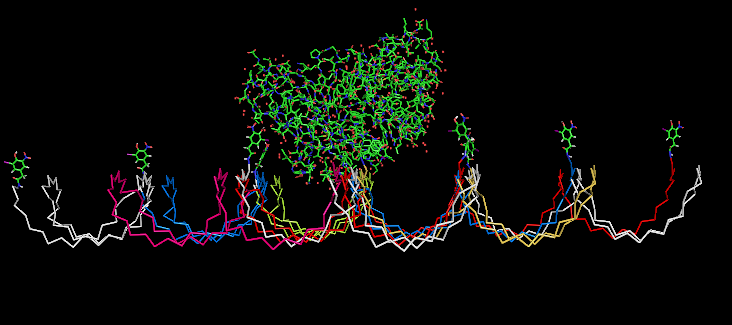 Fig.5: Oligo-pattern with NIP-molecules and Fab-fragment, binding accessibility estimation model
|
Fusion Proteins
The pictures below show models of the fusion proteins of our composite parts Bba_K157032 and Bba_K157033 code for (generated with "SwissPdbViewer"):
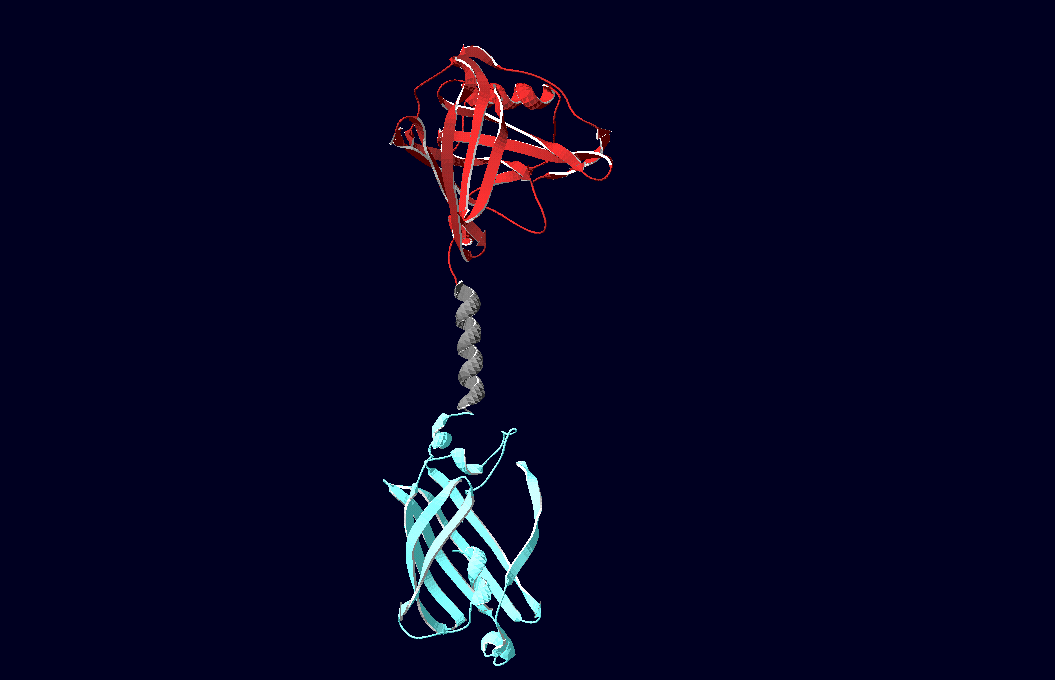 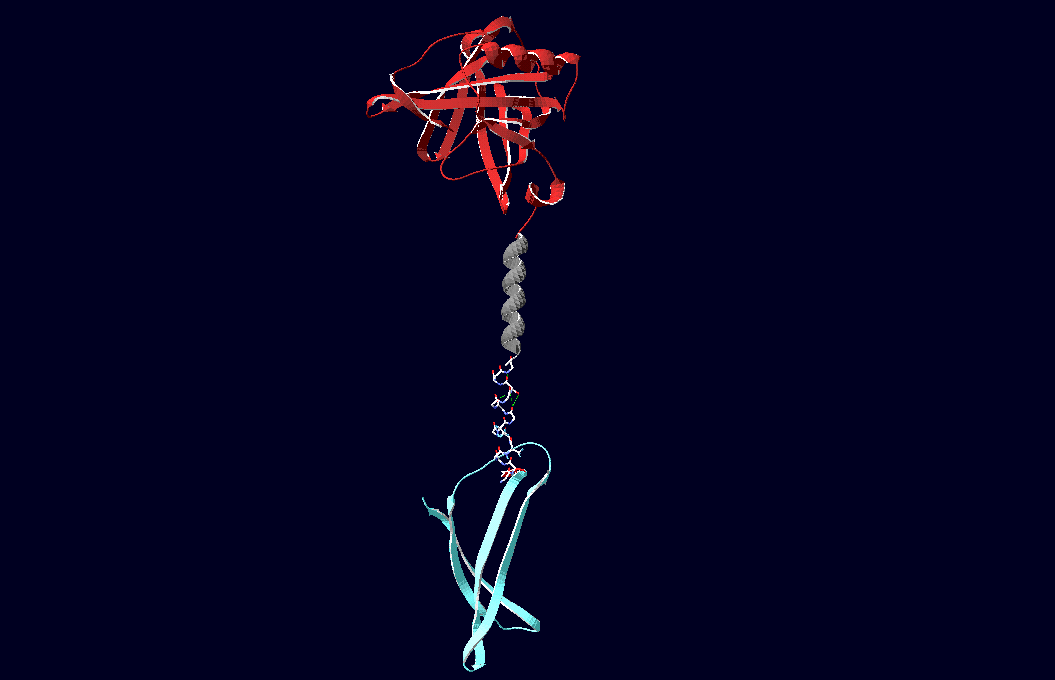
Fig.6: Red: LipocalinFluA, Grey: "Transmembrane region (BCR, no structural data; symbolized by helix), Blue: left: Split-Cerulean (CFP), N-terminal fragment; right: Split-Cerulean (CFP), C-terminal fragment
Following pdb-files have been used to generate the models:
Cerulean-CFP: 2Q57
Venus-YFP: 1MYW
Engineered Lipocalin: 1n0s
B1-8 Fv-Fragment:1a6w
|

 "
"







