Team:Waterloo/Project
From 2008.igem.org
(Difference between revisions)
m (added john's design diagram) |
|||
| Line 19: | Line 19: | ||
[[Image:2008Design.png]] | [[Image:2008Design.png]] | ||
| - | == | + | == Construction Strategy == |
| - | + | ||
| + | *Already have RFP w/ RBS and TT - should be able to just stick pLac (e .g.) in front of this for preliminary verification. GFP may be a better option for later testing of expression post-kill due to its faster turnover giving better temporal resolution. | ||
| + | *If we did this, we could also use this (in a lacIq background, maybe?) as a preliminary form of control, WT (or lacIq) cell + reporter plasmid (pLac+rfp) | ||
| + | **Test reporter by IPTG induction | ||
| + | **WT (or lacIq) cell + reporter plasmid with nuclease operon (e. g. under TetR control- have some semi-constructed stuff for this, tight enough control for growth?) | ||
| + | **Test for reporter expression upon expression of nuclease (aTe derepression of pTet-controlled nuclease operon) | ||
| + | *Later, could have quorum sensing control T7PoI expression (q.s. + T7 stuff may already exist together in the registry?), and put the nuclease operon under T7 control (instead of TetR) | ||
| Line 37: | Line 42: | ||
=== Part 3 === | === Part 3 === | ||
| - | |||
| - | |||
| - | |||
== Results == | == Results == | ||
Revision as of 02:36, 11 July 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Contents |
Overall project
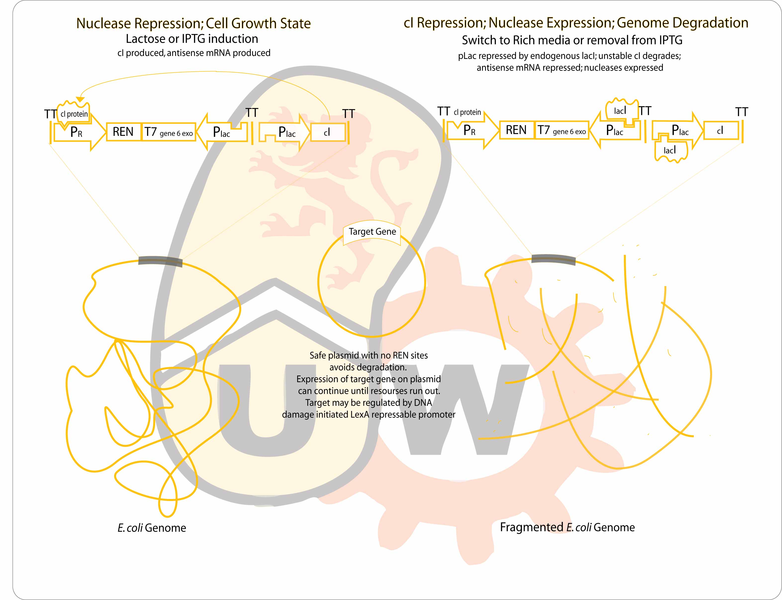
Our goal is to engineer a genome-free, cell-based expression system capable of producing a desired protein in response to environmental signals. The genome will be degraded by the combined activity of a restriction endonuclease (to fragment the genome) and an exonuclease (to hasten degradation of the genome). The gene for the protein of interest will be located on a plasmid which will lack recognition sites for the endonuclease, enabling it to remain intact after genome degradation. Expression of plasmid genes is expected to continue for a period of time until the "cell" expires.
The primary application of this would be an in situ compound production and delivery system for agricultural and/or therapeutic uses.
Construction Strategy
- Already have RFP w/ RBS and TT - should be able to just stick pLac (e .g.) in front of this for preliminary verification. GFP may be a better option for later testing of expression post-kill due to its faster turnover giving better temporal resolution.
- If we did this, we could also use this (in a lacIq background, maybe?) as a preliminary form of control, WT (or lacIq) cell + reporter plasmid (pLac+rfp)
- Test reporter by IPTG induction
- WT (or lacIq) cell + reporter plasmid with nuclease operon (e. g. under TetR control- have some semi-constructed stuff for this, tight enough control for growth?)
- Test for reporter expression upon expression of nuclease (aTe derepression of pTet-controlled nuclease operon)
- Later, could have quorum sensing control T7PoI expression (q.s. + T7 stuff may already exist together in the registry?), and put the nuclease operon under T7 control (instead of TetR)
 "
"