Team:KULeuven/Model/Cell Death
From 2008.igem.org
(→Describing the system) |
|||
| Line 140: | Line 140: | ||
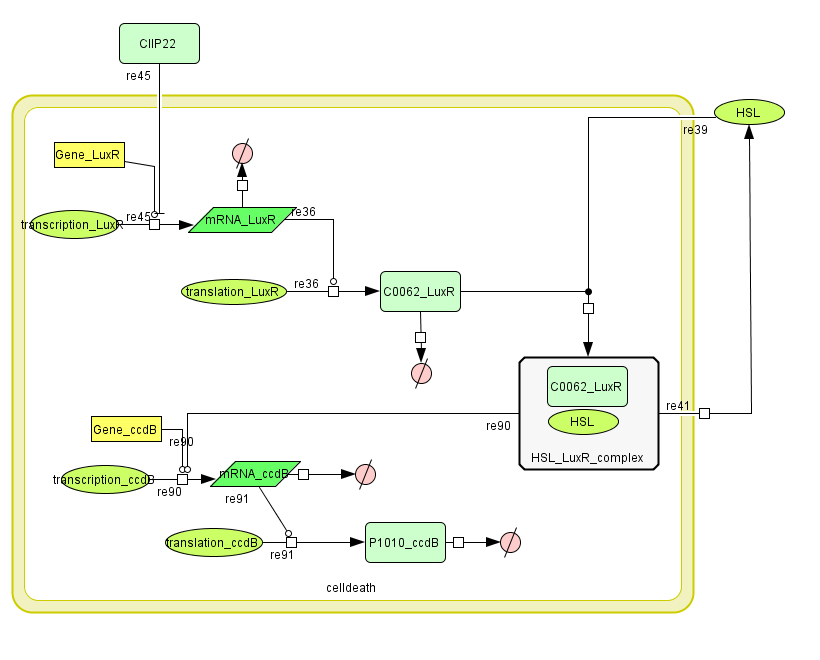
==== CellDesigner ([https://static.igem.org/mediawiki/2008/3/37/CellDeath_CellDesigner.zip SBML file])==== | ==== CellDesigner ([https://static.igem.org/mediawiki/2008/3/37/CellDeath_CellDesigner.zip SBML file])==== | ||
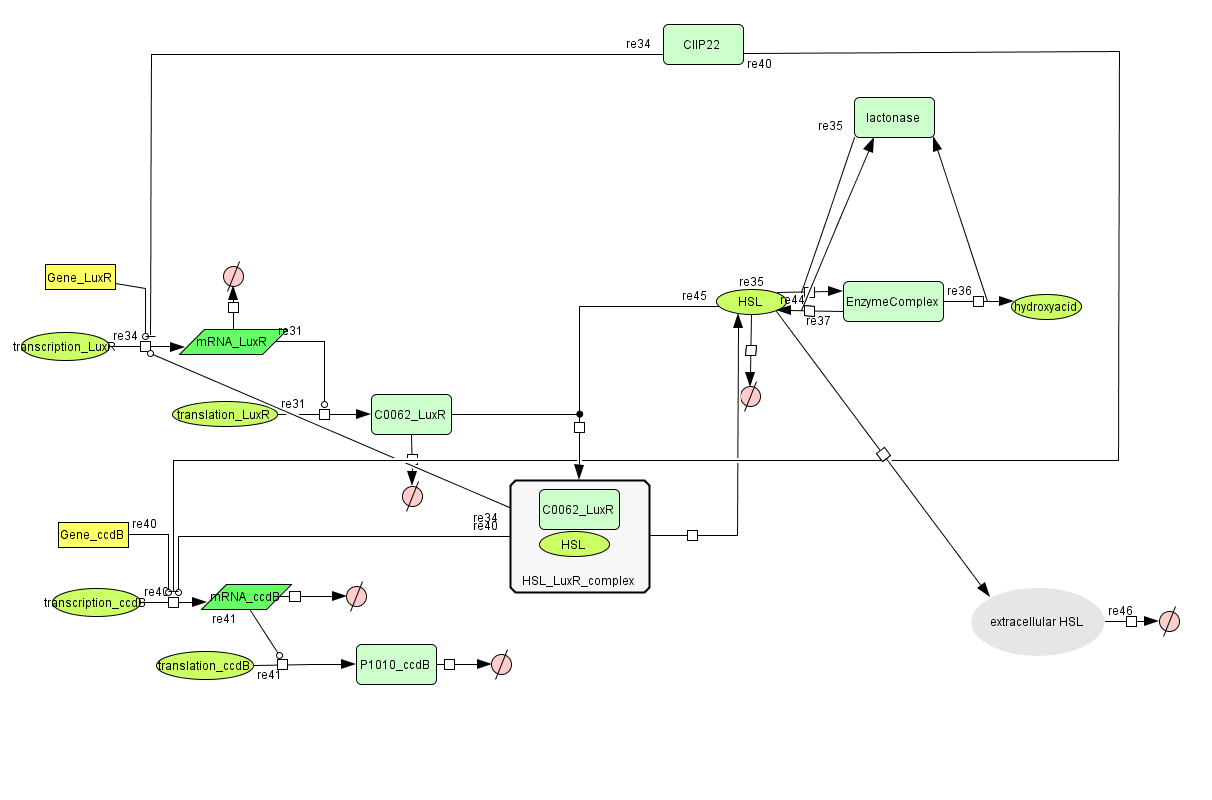
| - | [[Image:CellDeath_CellDesigner.png| | + | [[Image:CellDeath_CellDesigner.png|600px|center|Cell Death]] |
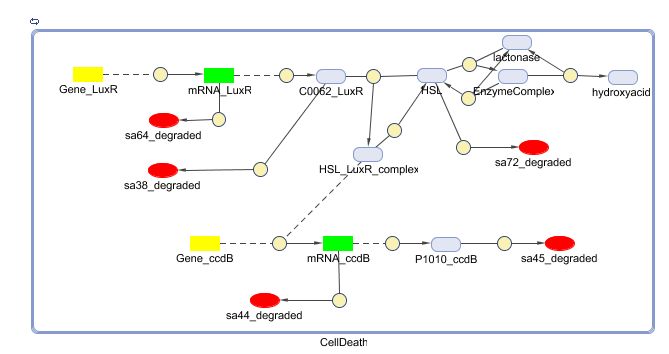
==== Matlab ([https://static.igem.org/mediawiki/2008/4/45/CellDeath_Matlab.zip SBML file])==== | ==== Matlab ([https://static.igem.org/mediawiki/2008/4/45/CellDeath_Matlab.zip SBML file])==== | ||
Remark: not yet up to date to latest (final) version | Remark: not yet up to date to latest (final) version | ||
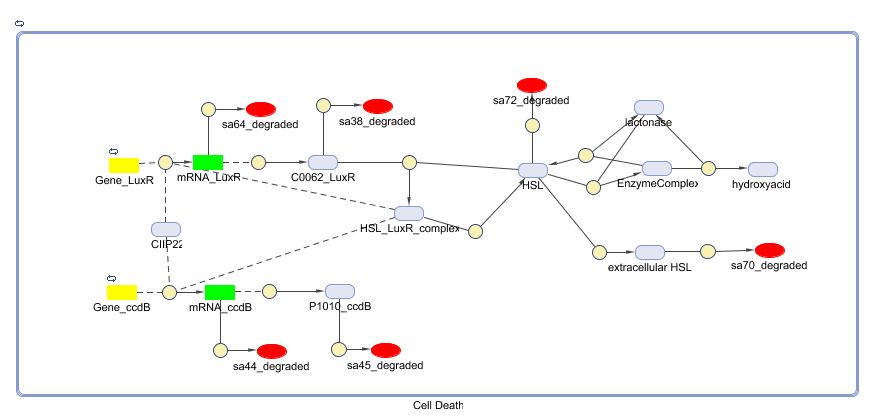
| - | [[Image:CellDeath_Matlab.jpg| | + | [[Image:CellDeath_Matlab.jpg|600px|center|Cell Death]] |
=== Simulations === | === Simulations === | ||
| - | todo | + | todo!!! |
=== Sensitivity Analysis === | === Sensitivity Analysis === | ||
| - | todo | + | todo!!! |
== New Cell Death== | == New Cell Death== | ||
| Line 161: | Line 161: | ||
====ODE's==== | ====ODE's==== | ||
| - | ==== | + | |
| + | todo!!!<br> | ||
| + | |||
| + | ==== Parameters ==== | ||
{| width=80% style="border: 1px solid #003E81; background-color: #EEFFFF;" | {| width=80% style="border: 1px solid #003E81; background-color: #EEFFFF;" | ||
|+ ''Parameter values (Cell Death)'' | |+ ''Parameter values (Cell Death)'' | ||
| Line 269: | Line 272: | ||
[[Image:CellDeath_Matlab_Final.jpg|800px|center|Cell Death]] | [[Image:CellDeath_Matlab_Final.jpg|800px|center|Cell Death]] | ||
| + | |||
| + | === Simulations === | ||
| + | |||
| + | todo!!!<br> | ||
| + | |||
| + | === Sensitivity Analysis === | ||
| + | |||
| + | todo!!! | ||
Revision as of 09:50, 26 August 2008
Contents |
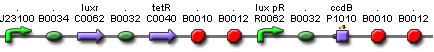
Cell Death
Position in the system
The Cell Death subsystem receives input from two other subsystems, namely:
LuxR is the component repressing the regulation of CcdB, the toxic product causing cell death. There the LuxR production is constitutive, no protein controls the gene regulation of LuxR, but the amount of LuxR available to repress the transcripion of the CcdB gene is controlled by HSL (Homoserine lactone).
If the inverter subsystem produces HSL (occurs when no light is detectable), this will forms a complex with LuxR. This will diminish the amount of LuxR available to repress the CcdB transcription and initiate cell death. When waiting long enough the amount of HSL becomes critical.
If however the pulse generator becomes active (by the filter), it will produce a pulse of lactonase, which will then bind to the HSL, reacting to an hydroxy-acid. As opposed to HSL, this hydroxy-acid will no longer form a complex with LuxR. This increase in LuxR lowers the CcdB production. The challenge is to generate a pulse of lactonase high enough to neutralise all HSL present in the cell.
Describing the system
ODE's
Parameters
Remark: update parameters to repressive promotor
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dLuxR | 0.0010 s-1 | ||
| dLuxR_HSL | 0.0010 s-1 | complex of HSL and LuxR degrades, giving back HSL | |
| dRNA_LuxR | 0.00227 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dCcdB | 7.7E-5 s-1 | [http://www.ncbi.nlm.nih.gov/pubmed/8022284?dopt=abstract link] | |
| dRNA_CcdB | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dHSL | 1.02E-6 s-1 | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] | |
| Association/Dissociation/Reaction Rates | |||
| kass (HSL+LuxR) | 1E6 s-1 | association rate of HSL with LuxR. Chosen to be relatively (to the other rate constants) high and such that Kdiss (HSL + LuxR) equals 10-6 | |
| kdiss (HSL+LuxR) | 1 s-1 | dissociation rate of the HSL-LuxR complex | |
| kass (HSL+lactonase) | 1E6 s-1 | association rate of HSL with lactonase | |
| kdiss (HSL+lactonase) | 446.5 s-1 | dissociation rate of the HSL-lactonase complex | |
| kcat (HSL:hydroxy-acid) | 29 s-1 | catalytic transformation of HSL to an hydroxy-acid, lactonase is the enzyme | [http://www.jbc.org/cgi/reprint/M311194200v1.pdf link] |
| Dissociation Constants | |||
| KHSL_LuxR | 1E-6 [M]/L | kdiss / kass (HSL+LuxR) | [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| KHSL_LuxR | 4.05E-6 [M]/L | binding HSL_LuxR on LuxPromotor | [http://parts.mit.edu/igem07/index.php/Tokyo/AHL_assay link] |
| Hill Cooperativity | |||
| nHSL_LuxR | 2.08 | [http://parts.mit.edu/igem07/index.php/Tokyo/AHL_assay link] | |
| Transcription Rates | |||
| kmRNA_LuxR (constitutive promotor) | 0.025 s-1 | see Constitutive Promotors & E. coli transcription Rates | |
| kmRNA_CcdB | 0.025 s-1 | maximal transcription rate for CcdB RNA (no LuxR repressor present) | |
[http://www.sciencedirect.com/science?_ob=MImg&_imagekey=B6T39-3Y6HKD6-BK-1&_cdi=4941&_user=877992&_orig=search&_coverDate=08%2F30%2F1995&_sk=998379998&view=c&wchp=dGLbVtz-zSkzS&md5=49efd14150c71e668eabdef225220ce3&ie=/sdarticle.pdf This paper] says that synthesis of even a few molecules of the shorter CcdB protein is probably lethal.
Models
CellDesigner (SBML file)
Matlab (SBML file)
Remark: not yet up to date to latest (final) version
Simulations
todo!!!
Sensitivity Analysis
todo!!!
New Cell Death
Describing the system
ODE's
todo!!!
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dLuxR | 0.0010 s-1 | ||
| dLuxR_HSL | 0.0010 s-1 | complex of HSL and LuxR degrades, giving back HSL | |
| dRNA_LuxR | 0.00227 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dCcdB | 7.7E-5 s-1 | [http://www.ncbi.nlm.nih.gov/pubmed/8022284?dopt=abstract link] | |
| dRNA_CcdB | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dHSL | 1.02E-6 s-1 | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] | |
| Association/Dissociation/Reaction Rates | |||
| kass (HSL+LuxR) | 1E6 s-1 | association rate of HSL with LuxR. Chosen to be relatively (to the other rate constants) high and such that Kdiss (HSL + LuxR) equals 10-6 | |
| kdiss (HSL+LuxR) | 1 s-1 | dissociation rate of the HSL-LuxR complex | |
| kass (HSL+lactonase) | 1E6 s-1 | association rate of HSL with lactonase | |
| kdiss (HSL+lactonase) | 446.5 s-1 | dissociation rate of the HSL-lactonase complex | |
| kcat (HSL:hydroxy-acid) | 29 s-1 | catalytic transformation of HSL to an hydroxy-acid, lactonase is the enzyme | [http://www.jbc.org/cgi/reprint/M311194200v1.pdf link] |
| Dissociation Constants | |||
| KHSL_LuxR | 1E-6 [M]/L | kdiss / kass (HSL+LuxR) | [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| KHSL_LuxR | 4.05E-6 [M]/L | binding HSL_LuxR on LuxPromotor | [http://parts.mit.edu/igem07/index.php/Tokyo/AHL_assay link] |
| Hill Cooperativity | |||
| nHSL_LuxR | 2.08 | [http://parts.mit.edu/igem07/index.php/Tokyo/AHL_assay link] | |
| Transcription Rates | |||
| kmRNA_LuxR (constitutive promotor) | 0.025 s-1 | see Constitutive Promotors & E. coli transcription Rates | |
| kmRNA_CcdB | 0.025 s-1 | maximal transcription rate for CcdB RNA (no LuxR repressor present) | |
Models
CellDesigner (SBML file)
Matlab (SBML file)
Simulations
todo!!!
Sensitivity Analysis
todo!!!
 "
"