Team:KULeuven/Model/Inverter
From 2008.igem.org
(Difference between revisions)
(→Describing the system) |
(→Parameters) |
||
| Line 37: | Line 37: | ||
|- | |- | ||
| d<sub>LuxI</sub> | | d<sub>LuxI</sub> | ||
| - | | | + | | d<sub>LVA</sub> = 2.814E-4 s<sup>-1</sup> |
| - | | | + | | LVA-tag reduces lifetime to 40 minutes |
| - | | | + | | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=106306 link] |
|- | |- | ||
| d<sub>RNA_LuxI</sub> | | d<sub>RNA_LuxI</sub> | ||
| Line 47: | Line 47: | ||
|- | |- | ||
| d<sub>LuxI_antimRNA</sub> | | d<sub>LuxI_antimRNA</sub> | ||
| - | | 0. | + | | 0.0045303737 s<sup>-1</sup> |
| - | | | + | | estimate: because this RNA isn't translated, it degrades faster |
| | | | ||
|- | |- | ||
| d<sub>LacI</sub> | | d<sub>LacI</sub> | ||
| - | | | + | | d<sub>LVA</sub> = 2.814E-4 s<sup>-1</sup> |
| - | | | + | | LVA-tag reduces lifetime to 40 minutes |
| - | | [http:// | + | | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=106306 link] |
|- | |- | ||
| - | | d<sub>LacI</sub> | + | | d<sub>closed mRNA LacI</sub> |
| 0.0046209812 s<sup>-1</sup> | | 0.0046209812 s<sup>-1</sup> | ||
| estimate: because this mRNA isn't translated, it degrades faster | | estimate: because this mRNA isn't translated, it degrades faster | ||
| | | | ||
|- | |- | ||
| - | | d<sub>LacI</sub> | + | | d<sub>open mRNA LacI</sub> |
| 0.0023104906 s<sup>-1</sup> | | 0.0023104906 s<sup>-1</sup> | ||
| | | | ||
| [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | ||
|- | |- | ||
| - | | d<sub> | + | | d<sub>open mRNA LacI complex</sub> |
| 0.0023104906 s<sup>-1</sup> | | 0.0023104906 s<sup>-1</sup> | ||
| | | | ||
| Line 73: | Line 73: | ||
| d<sub>HSL</sub> | | d<sub>HSL</sub> | ||
| 1.02E-6 s<sup>-1</sup> | | 1.02E-6 s<sup>-1</sup> | ||
| - | | | + | | very stable in the medium, lifetime around 185h |
| [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] | | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] | ||
|- | |- | ||
| Line 82: | Line 82: | ||
| Complex of LuxI mRNA with antisense mRNA | | Complex of LuxI mRNA with antisense mRNA | ||
| | | | ||
| + | |- | ||
| + | ! colspan="4" style="border-bottom: 1px solid #003E81;" | T7 Transcription | ||
| + | |- | ||
| + | | K<sub>T7</sub> | ||
| + | | 421 | ||
| + | | dissociation constant, recalculated to remove units | ||
| + | | [http://www.jbc.org/cgi/content/full/279/5/3239 link] | ||
| + | |- | ||
| + | | k<sub>max</sub> | ||
| + | | 0.044 s<sup>-1</sup> | ||
| + | | maximal T7 transcription rate | ||
| + | | [http://www.jbc.org/cgi/content/full/279/5/3239 link] | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Transcription Rates | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Transcription Rates | ||
| Line 95: | Line 107: | ||
| | | | ||
|- | |- | ||
| - | ! colspan="4" style="border-bottom: 1px solid #003E81;" | | + | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Transcription Repression |
|- | |- | ||
| - | | K<sub> | + | | K<sub>LacI</sub> M |
| - | | 1.0E-10 | + | | 1.0E-10 M<sup>-1</sup> |
| - | | | + | | Dissociation constant |
| | | | ||
|- | |- | ||
| - | | n<sub> | + | | n<sub>LacI</sub> |
| 2.0 | | 2.0 | ||
| - | | | + | | Hill coefficient for LacI |
| - | + | | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | [http:// | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Translation Rates | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Translation Rates | ||
| Line 121: | Line 123: | ||
| k<sub>LuxI</sub> | | k<sub>LuxI</sub> | ||
| 0.167 s<sup>-1</sup> | | 0.167 s<sup>-1</sup> | ||
| - | | | + | | translation rate for B0032 RBS (0.3 relative efficiency) |
| | | | ||
|- | |- | ||
| k<sub>LacI</sub> | | k<sub>LacI</sub> | ||
| 0.167 s<sup>-1</sup> | | 0.167 s<sup>-1</sup> | ||
| - | | RBS is situated in the keylock | + | | estimate: RBS is situated in the keylock |
| | | | ||
|- | |- | ||
| Line 138: | Line 140: | ||
| K<sub>mRNA_LacI</sub> | | K<sub>mRNA_LacI</sub> | ||
| 0.015 | | 0.015 | ||
| - | | | + | | between closed and open LacI mRNA, experimental |
| - | | | + | | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
|- | |- | ||
| K<sub>mRNA_LacI_Key</sub> | | K<sub>mRNA_LacI_Key</sub> | ||
| 0.0212 /M | | 0.0212 /M | ||
| - | | | + | | between closed LacI mRNA and key unlocked mRNA complex, experimental |
| - | | | + | | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link]| |
| + | |- | ||
| + | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Rate constants | ||
| + | |- | ||
| + | | k<sub>dis2</sub> | ||
| + | | 100 s<sup>-1</sup> | ||
| + | | derived from experimental values | ||
| + | | | ||
| + | |- | ||
| + | | k<sub>complex2</sub> | ||
| + | | 57 s<sup>-1</sup> | ||
| + | | derived from experimental values | ||
| + | | | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Reaction Rates | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Reaction Rates | ||
Revision as of 15:00, 27 August 2008
dock/undock dropdown
Contents |
Invertimer
Position in the system
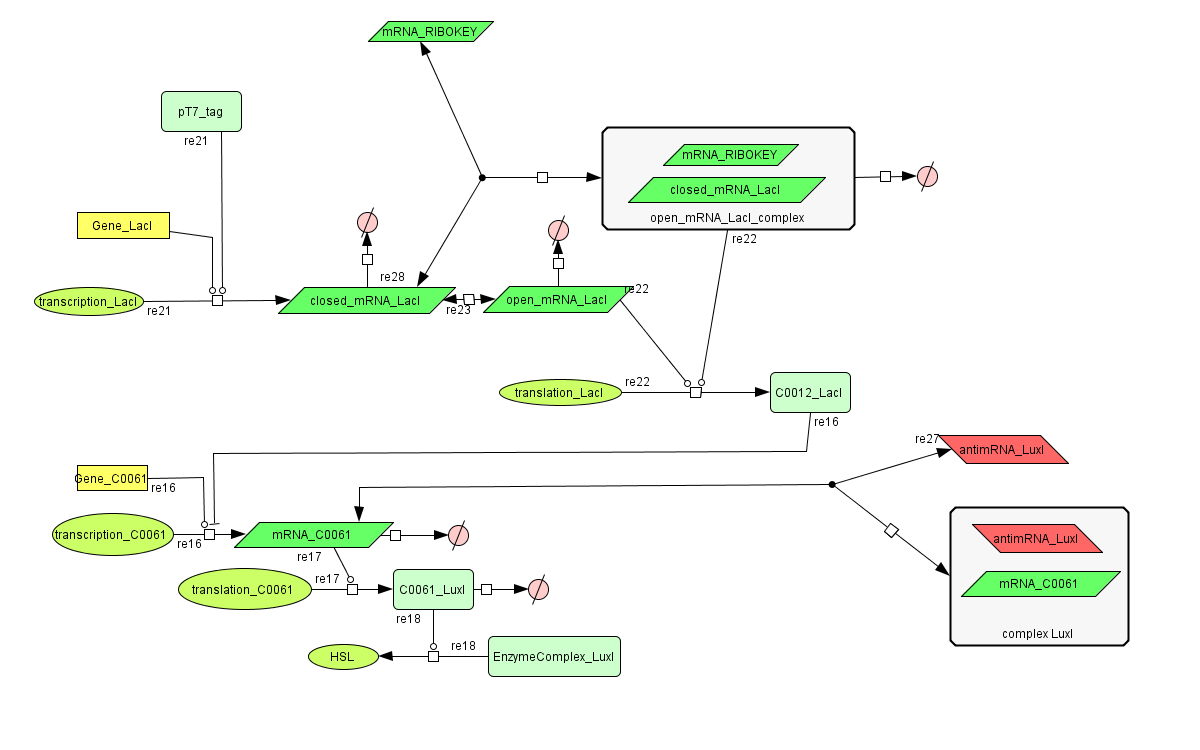
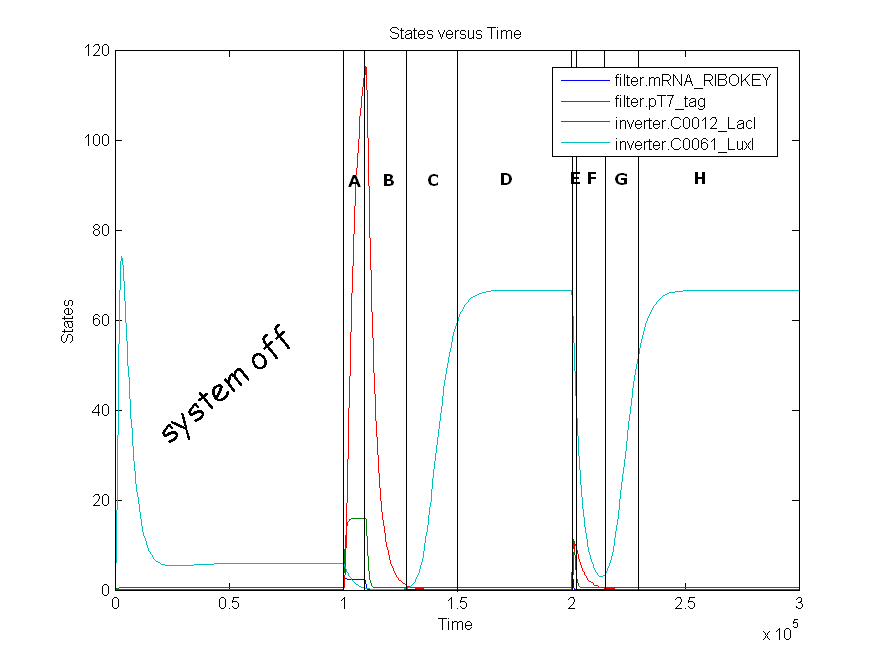
The invertimer subsystem receives its input from the filter, T7. The invertimer's function is to produce HSL when no input is present, so a low T7 input gives rise to a high HSL output and vice versa. The production of HSL means that the cell will start a timer that eventually will be used in the celldeath-subsystem to produce ccdB. In this way the cell will die off if no desease remains present.
Describing the system
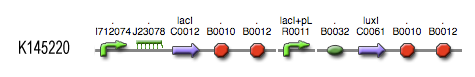
see also: Project:Invertimer
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dLuxI | dLVA = 2.814E-4 s-1 | LVA-tag reduces lifetime to 40 minutes | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=106306 link] |
| dRNA_LuxI | 0.0025 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dLuxI_antimRNA | 0.0045303737 s-1 | estimate: because this RNA isn't translated, it degrades faster | |
| dLacI | dLVA = 2.814E-4 s-1 | LVA-tag reduces lifetime to 40 minutes | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=106306 link] |
| dclosed mRNA LacI | 0.0046209812 s-1 | estimate: because this mRNA isn't translated, it degrades faster | |
| dopen mRNA LacI | 0.0023104906 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen mRNA LacI complex | 0.0023104906 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dHSL | 1.02E-6 s-1 | very stable in the medium, lifetime around 185h | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] |
| Dissociation Rates | |||
| KmRNA_LuxI:antisense_mRNA | 4.22E14 | Complex of LuxI mRNA with antisense mRNA | |
| T7 Transcription | |||
| KT7 | 421 | dissociation constant, recalculated to remove units | [http://www.jbc.org/cgi/content/full/279/5/3239 link] |
| kmax | 0.044 s-1 | maximal T7 transcription rate | [http://www.jbc.org/cgi/content/full/279/5/3239 link] |
| Transcription Rates | |||
| kmRNA_LacI | 0.0011 s-1 | ||
| kmRNA_LuxI | 0.0025 s-1 | ||
| Transcription Repression | |||
| KLacI M | 1.0E-10 M-1 | Dissociation constant | |
| nLacI | 2.0 | Hill coefficient for LacI | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] |
| Translation Rates | |||
| kLuxI | 0.167 s-1 | translation rate for B0032 RBS (0.3 relative efficiency) | |
| kLacI | 0.167 s-1 | estimate: RBS is situated in the keylock | |
| kT7 | 182 s-1 | ||
| Equilibrium Constants | |||
| KmRNA_LacI | 0.015 | between closed and open LacI mRNA, experimental | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| KmRNA_LacI_Key | 0.0212 /M | between closed LacI mRNA and key unlocked mRNA complex, experimental | |
| Rate constants | |||
| kdis2 | 100 s-1 | derived from experimental values | |
| kcomplex2 | 57 s-1 | derived from experimental values | |
| Reaction Rates | |||
| kLuxI:HSL | 0.0167 s-1 | [http://www.pnas.org/content/93/18/9505.full.pdf+html link Vmax] | |
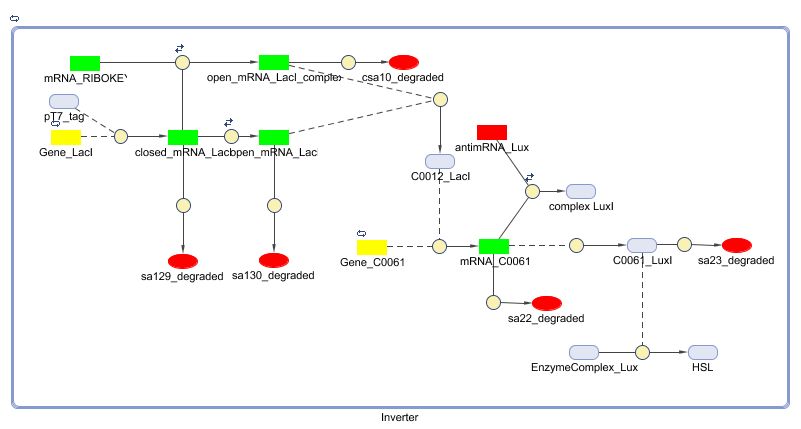
Models
CellDesigner (SBML file)
Matlab (SBML file)
Remark: not yet up to date to latest (final) version
Simulations
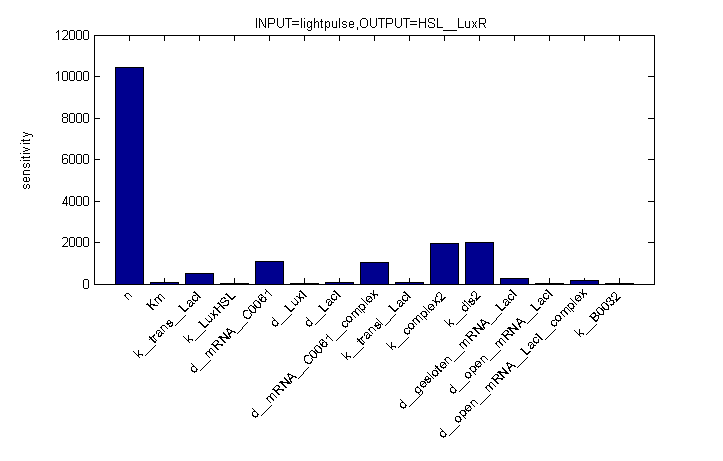
Remark: simulation is not from latest version.
 "
"