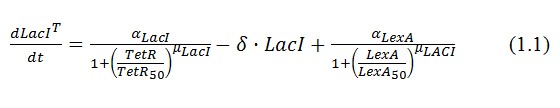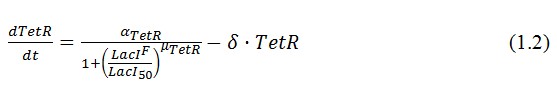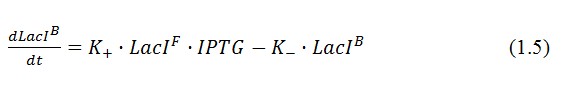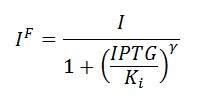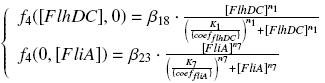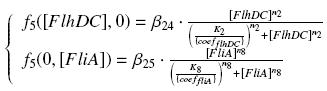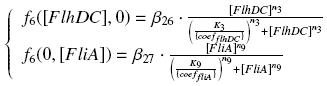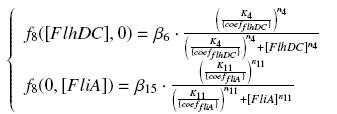Team:Bologna/Modeling
From 2008.igem.org
(Difference between revisions)
(→Mathematical Model) |
(→Mathematical Model) |
||
| Line 27: | Line 27: | ||
[[Image:Circuito2.jpg|700px|center]] | [[Image:Circuito2.jpg|700px|center]] | ||
| - | The | + | The genetic circuit in Figure 1 can be modeled with the following equations: |
<br><br> | <br><br> | ||
Revision as of 09:14, 16 October 2008
| HOME | THE PROJECT | THE TEAM | PARTS SUBMITTED TO THE REGISTRY | MODELING | NOTEBOOK | BIOSAFETY |
|---|
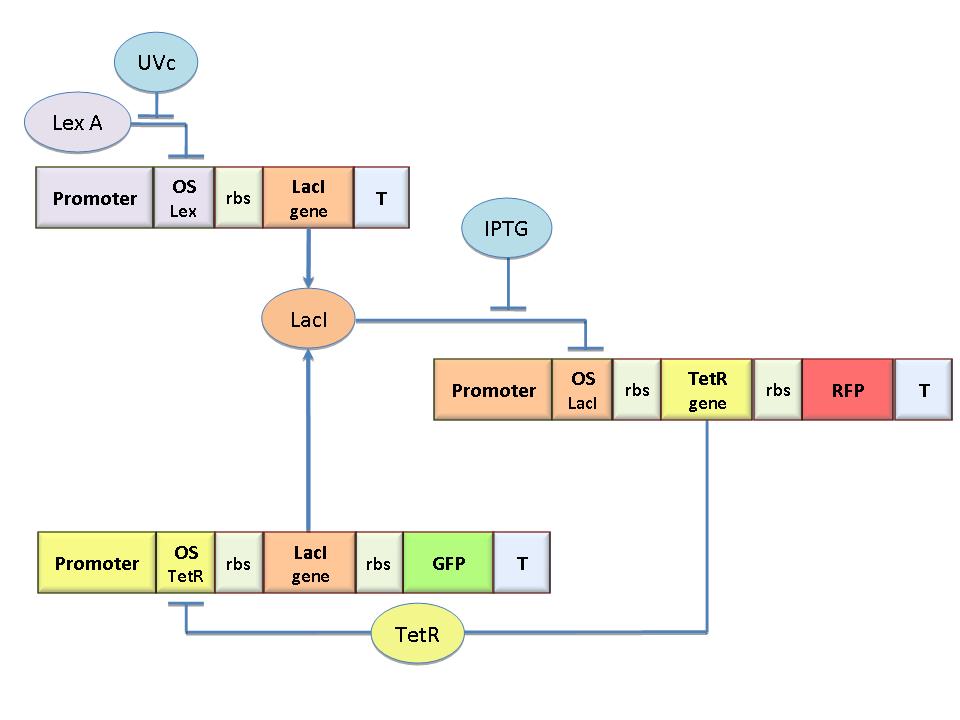
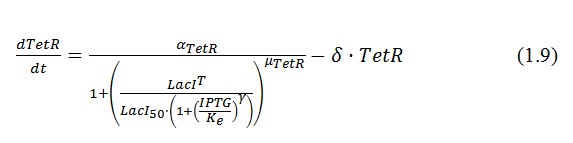
Mathematical Model
The genetic circuit in Figure 1 can be modeled with the following equations:
Where:
α = promoter activity and rbs
δ = coefficient of degradation (cellular diluition)
μ = coefficient of cooperativity
The circuit in Figure 1 can be modeled with the following equations:
Where:
In the model we distinguish between protein binded to repressor ()and protein free, not binded to repressor (), in this case repressor is represented by IPTG. Knowing that and the law of mass action is possible write where we can replace represents the entry of IPTG inside the cell. The same thing can be done even for LexA obtaining where represents the UV radiation.Placing:
The dimensionless equations are:
Hypothesizing: to be under conditions of equilibrium;
all the entries are void (); the affinity of the repressor for the operator site is high, for as we have been defined it will surely be smaller of one so ;the cooperativity is the same both for the LacI and for TetR and it is worth 2; the equations become:
 "
"