Team:UC Berkeley/GatewayOverview
From 2008.igem.org
(→Gateway Chemistry) |
(→Gateway Chemistry) |
||
| Line 24: | Line 24: | ||
[[Image:LR reaction.jpg|frame|center|Gateway LR reaction where gene of interest (flanked by attL sites) is transferred to destination vector containing attR sites. <br> Image source: http://www.bio.davidson.edu/courses/Molbio/MolStudents/spring2000/patton/gateway.htm]] | [[Image:LR reaction.jpg|frame|center|Gateway LR reaction where gene of interest (flanked by attL sites) is transferred to destination vector containing attR sites. <br> Image source: http://www.bio.davidson.edu/courses/Molbio/MolStudents/spring2000/patton/gateway.htm]] | ||
| + | * methods of selection | ||
<html> | <html> | ||
Revision as of 01:13, 28 October 2008
Contents |
Why Use Gateway?
The first step of the layered assembly scheme involves the transfer of biobrick parts from an entry vector to a double antibiotic assembly vector. Traditionally, this would require a fairly work-intensive protocol requiring digestion, gel purification, ligation, transformation, and plasmid isolation. In addition to being more time-consuming, the aforementioned procedure is also suboptimal because it is difficult to scale-up.
The [http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Cloning/Gateway-Cloning.html| Gateway Cloning] approach developed by Invitrogen offers a more efficient and convenient alternative for parts transfer. Their procedure involves the enzyme-catalyzed exchange of parts flanked by specific recombination sites. Experimentally, it is a one-pot, room-temperature reaction where the plasmids, buffer, water and enzymes are added together. After the addition of another enzyme and a short incubation period to terminate the reaction, the entire mixture can be transformed directly. This one-pot approach with a fewer steps is much more suitable for large-scale experimentation.
- traditionally used to go from one entry vector to multiple assembly vectors
Gateway Chemistry
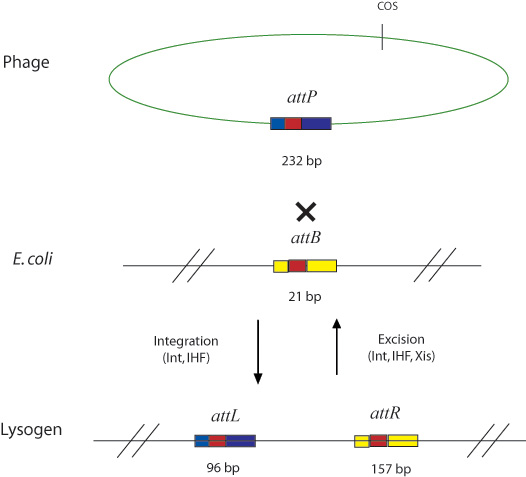
In general, Gateway reactions involve the attB, attP, attL, and attR recombination sites and the integrase (Int), excisionase (Xis), and integration host factor (IHF) enzymes. Invitrogen adapted these components from the original lambda phage recombination system shown below.
- brief description of what each enzyme does
In the Invitrogen scheme, the recombination sites are found in pairs flanking sequences that are intended for transfer. The recombination pairs are directional and specificity is given by ten nucleotides in the core region of each site, as shown below.
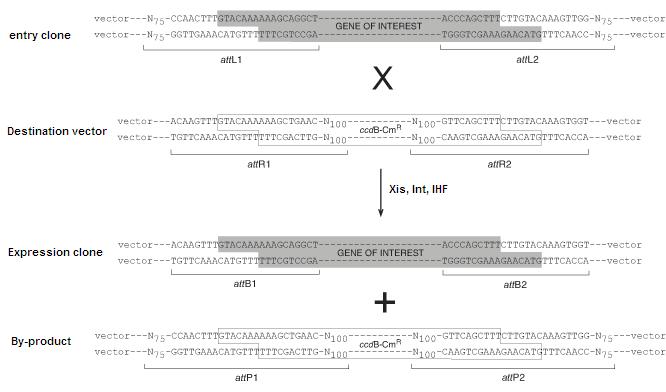
Image source:https://commerce.invitrogen.com/index.cfm?fuseaction=iProtocol.unitSectionTree&treeNodeId=290D0521B5FFA1B782C62C0AB62FD7BC
The schematic below depicts the LR reaction, during which the attL sites recombine with attR to yield attB and attP sites. The BP reaction proceeds in the opposite direction yielding attL and attR sites.
- methods of selection
Animation source: http://www.bio.davidson.edu/courses/Molbio/MolStudents/spring2000/patton/gateway.htm
Gateway in vivo
References
- http://www.bio.davidson.edu/courses/Molbio/MolStudents/spring2000/patton/gateway.htm
- http://tools.invitrogen.com/downloads/gateway-the-basics-seminar.html
- http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Cloning/Gateway-Cloning.html|Gateway
- https://commerce.invitrogen.com/index.cfm?fuseaction=iProtocol.unitSectionTree&treeNodeId=290D0521B5FFA1B782C62C0AB62FD7BC
 "
"