Team:UC Berkeley/LayeredAssembly
From 2008.igem.org
(→Project Motivation) |
|||
| Line 4: | Line 4: | ||
In an effort to assist in the automation of synthetic biology, we investigated methods to simplify liquid handling so that it can more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the ''E. coli'', thereby allowing them to perform the required protocols. | In an effort to assist in the automation of synthetic biology, we investigated methods to simplify liquid handling so that it can more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the ''E. coli'', thereby allowing them to perform the required protocols. | ||
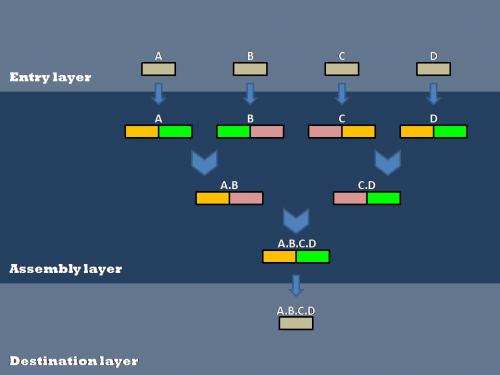
| - | Our efforts to optimize synthetic biology protocols centered around the scheme for layered assembly. This involves the creation of biobrick parts in an entry vector, their transfer into double antibiotic assembly vectors, and subsequent assembly steps to connect various parts. After all of the assembly steps are complete, the desired composite parts can then be transferred into the destination vector of choice. | + | Our efforts to optimize synthetic biology protocols centered around the scheme for layered assembly. This involves the creation of biobrick parts in an entry vector, their transfer into double antibiotic assembly vectors(antibiotic resistances shown in orange, green, and pink), and subsequent assembly steps to connect various parts(A, B, C, and D). After all of the assembly steps are complete, the desired composite parts can then be transferred into the destination vector of choice. |
[[Image:layered Assembly.jpg|center|frame|350px|A schematic representation of Layered Assembly]] | [[Image:layered Assembly.jpg|center|frame|350px|A schematic representation of Layered Assembly]] | ||
| + | their transfer into double antibiotic assembly vectors , and subsequent assembly steps to connect various parts | ||
Revision as of 01:15, 28 October 2008
Project Motivation
In an effort to assist in the automation of synthetic biology, we investigated methods to simplify liquid handling so that it can more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the E. coli, thereby allowing them to perform the required protocols.
Our efforts to optimize synthetic biology protocols centered around the scheme for layered assembly. This involves the creation of biobrick parts in an entry vector, their transfer into double antibiotic assembly vectors(antibiotic resistances shown in orange, green, and pink), and subsequent assembly steps to connect various parts(A, B, C, and D). After all of the assembly steps are complete, the desired composite parts can then be transferred into the destination vector of choice.
their transfer into double antibiotic assembly vectors , and subsequent assembly steps to connect various parts
 "
"