Team:Freiburg Cloning Strategy
From 2008.igem.org
(Difference between revisions)
| Line 3: | Line 3: | ||
<div style="font-size:18pt;"> | <div style="font-size:18pt;"> | ||
<font face="Arial Rounded MT Bold" style="color:#010369">_cloning strategy</font></div> | <font face="Arial Rounded MT Bold" style="color:#010369">_cloning strategy</font></div> | ||
| - | + | <br> | |
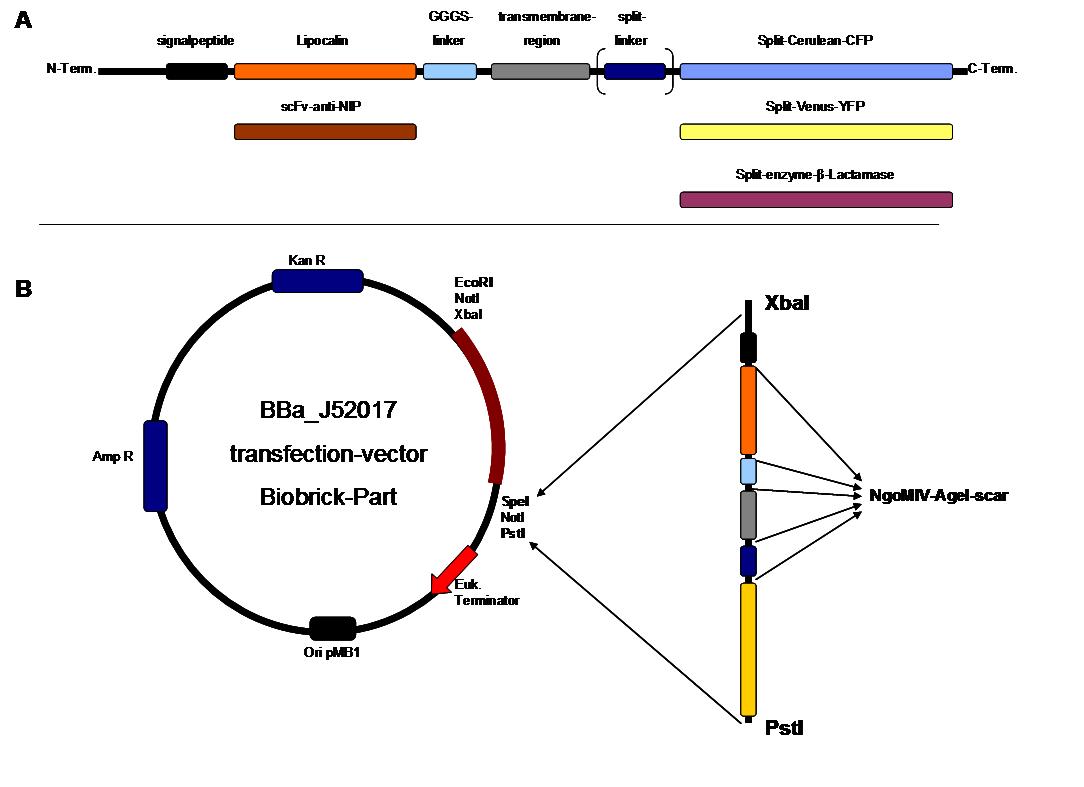
To form different types of synthetic receptor constructs a principle of a building set was used. There were two different forms of an extracellular binding protein and four different components of an intracellular signal transduction reporter protein, Cerulean CFP, Split Venus YFP, β-Lactamase and Luciferase. These reporters were used as Split-proteins to achieve that there is only detection when two receptors come together and form a cluster. With this strategy a signal is only given by two binding-molecules which are next to each other as it is realized in the DNA-origami structure or the NIP and Fluorescein coupled BSA. | To form different types of synthetic receptor constructs a principle of a building set was used. There were two different forms of an extracellular binding protein and four different components of an intracellular signal transduction reporter protein, Cerulean CFP, Split Venus YFP, β-Lactamase and Luciferase. These reporters were used as Split-proteins to achieve that there is only detection when two receptors come together and form a cluster. With this strategy a signal is only given by two binding-molecules which are next to each other as it is realized in the DNA-origami structure or the NIP and Fluorescein coupled BSA. | ||
The whole receptor construct consists out of a signalpeptide, an extracellular receptor domain, a GGGS-linker followed by transmembrane region and an intracellular split-reporter-protein. The signalpeptide-sequence according to the EGF-Receptor erbb1 ensures the transport of the protein construct to the cytoplasmamembrane. A GGGS-linker is necessary to create a distance between membrane and the recognition-site to overcome cell surface structures as glycoproteins and glyclipids. The sequence of the transmembrane region is appropriate to that of the EGF-Receptor erbb1 as well. The C-terminal split-parts of Venus-YFP and Cerulean CFP were not directly fused to the transmembrane-region. To get a little bit more flexibility in binding to the N-terminal part a Split Fluorophor-linker was assembled in between. | The whole receptor construct consists out of a signalpeptide, an extracellular receptor domain, a GGGS-linker followed by transmembrane region and an intracellular split-reporter-protein. The signalpeptide-sequence according to the EGF-Receptor erbb1 ensures the transport of the protein construct to the cytoplasmamembrane. A GGGS-linker is necessary to create a distance between membrane and the recognition-site to overcome cell surface structures as glycoproteins and glyclipids. The sequence of the transmembrane region is appropriate to that of the EGF-Receptor erbb1 as well. The C-terminal split-parts of Venus-YFP and Cerulean CFP were not directly fused to the transmembrane-region. To get a little bit more flexibility in binding to the N-terminal part a Split Fluorophor-linker was assembled in between. | ||
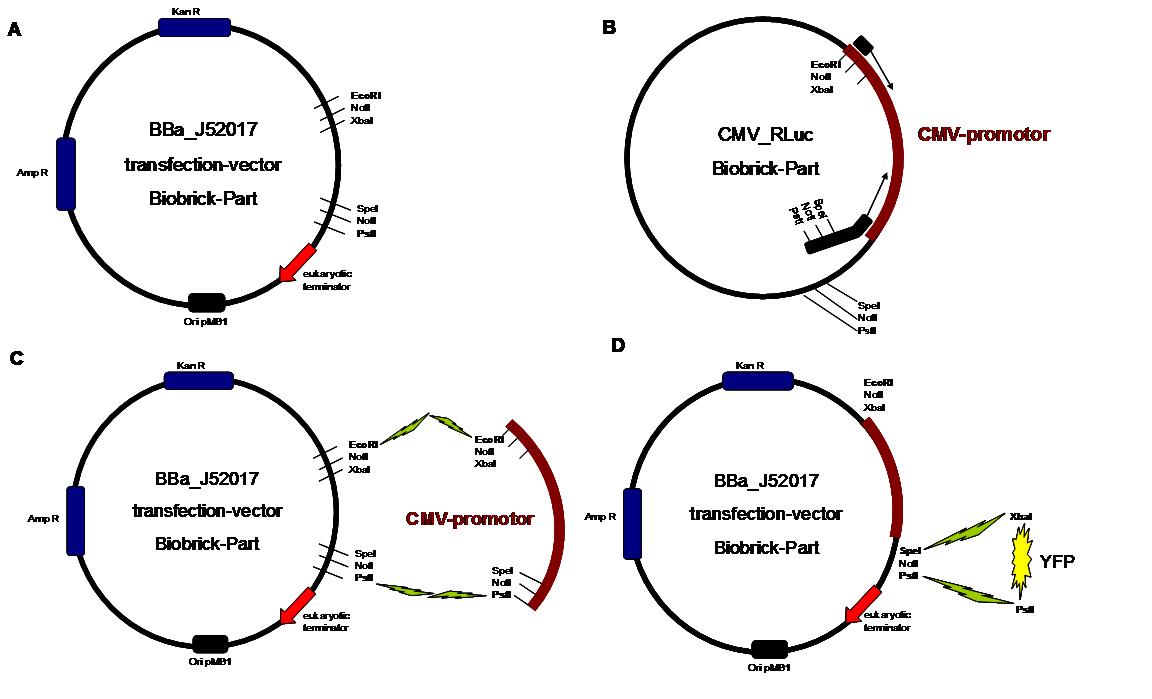
Almost all parts were ordered by genesynthesis in a pMA-vector system which is adapted for cloning BioBrick parts. All possible variations of the receptor constructs were fused together in the pMA-vector and afterwards inserted into a transfectionvector. | Almost all parts were ordered by genesynthesis in a pMA-vector system which is adapted for cloning BioBrick parts. All possible variations of the receptor constructs were fused together in the pMA-vector and afterwards inserted into a transfectionvector. | ||
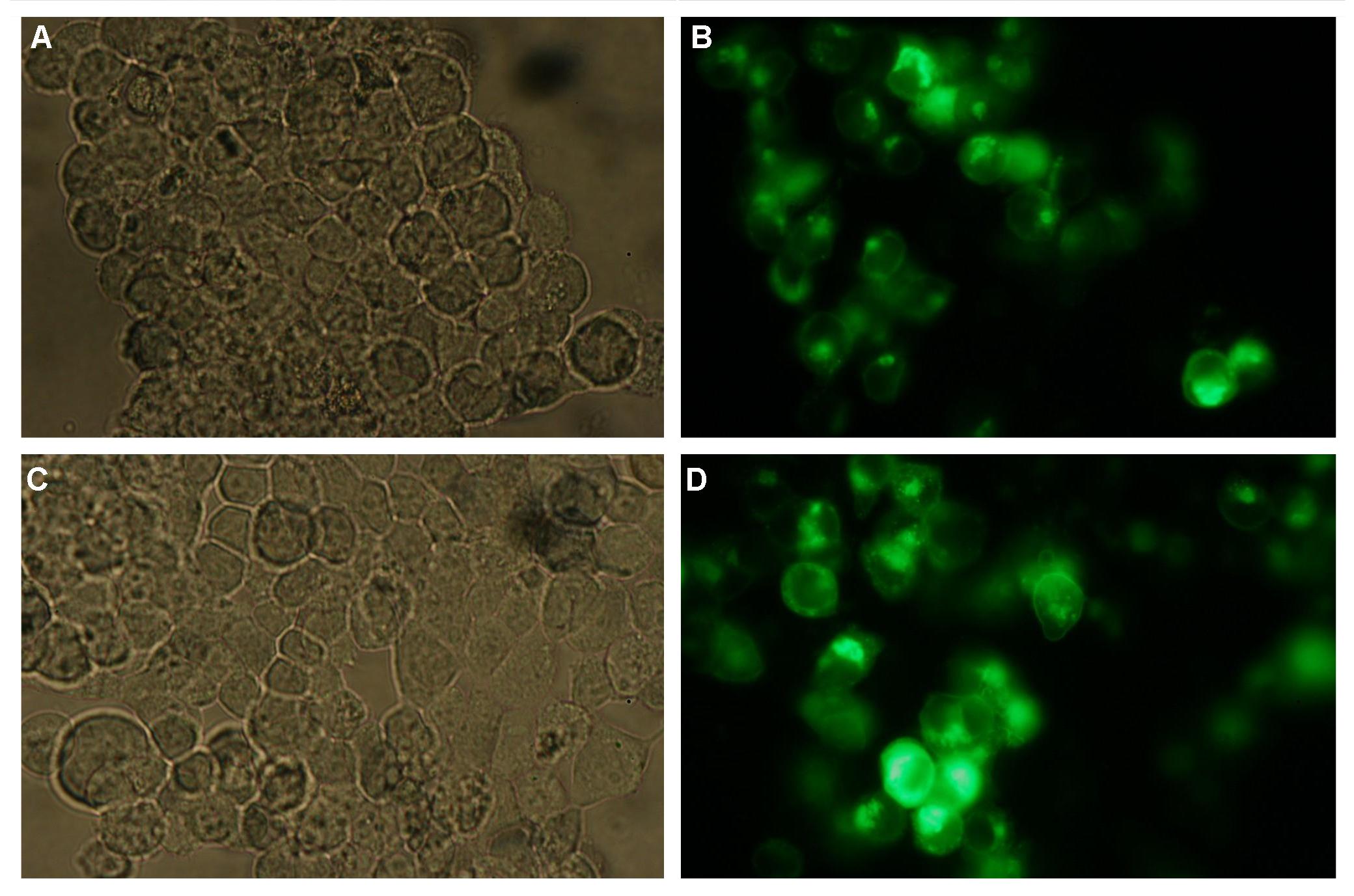
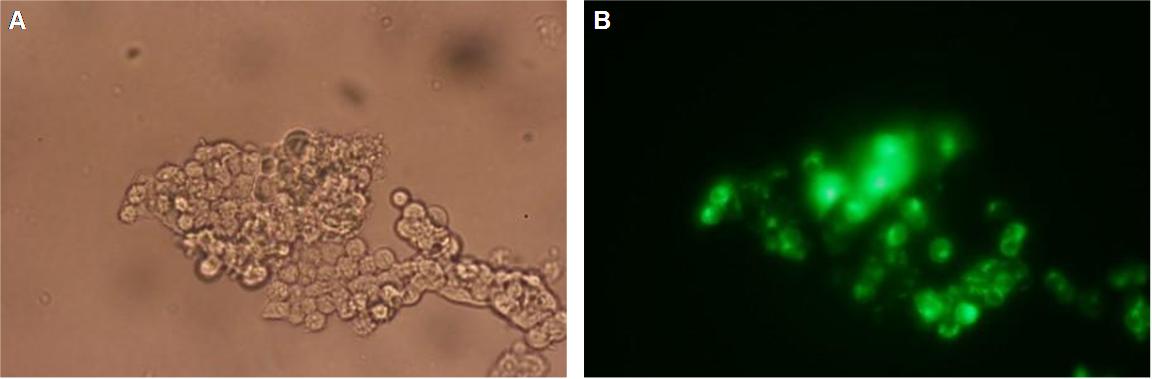
| - | The transfectionvector is a BioBrick 2007 part (BBa_J52017 http://partsregistry.org/ Part:BBa_J52017 ) and includes a kanamycin and ampicillin resistance cassette. There is a pMB1 Ori and the multiple cloning site consist out of the BioBrick Prefix and Suffix restriction enzymes (Prefix: EcoRI, NotI, XbaI/ Suffix: SpeI, NotI, PstI) followed by an eukaryotic terminator sequence. To achieve a strong expression a constitutive promoter construct was needed. Because of this Cauliflower Mosaik Virus [CMV] promotor was amplified by PCR using the BioBrick part BBa_J52038 (http://partsregistry.org/Part:BBa_J52038) and primers below-mentioned. The forward primer bound in the iGEM-prefix, while reverse primer bound at the end of the CMV-coding region. The reverse primer also contained an overhang to get the iGEM-suffix directly to the end of the promoter sequence which later allowed the cloning into the transfectionvector. CMV promoter was cloned into the vector by using the EcoRI and PstI restriction sites. To test the expression activity of the vector with CMV promoter the gene of the yellow fluorescent protein [YFP] was cloned behind the promoter region by using XbaI, PstI restriction enzymes for the YFP insert and SpeI, PstI for the vector to open Biobrick suffix. The tranfsfection of the resulting plasmid into 293T cells shows fully functionality.<br> | + | The transfectionvector is a BioBrick 2007 part (BBa_J52017 http://partsregistry.org/Part:BBa_J52017 ) and includes a kanamycin and ampicillin resistance cassette. There is a pMB1 Ori and the multiple cloning site consist out of the BioBrick Prefix and Suffix restriction enzymes (Prefix: EcoRI, NotI, XbaI/ Suffix: SpeI, NotI, PstI) followed by an eukaryotic terminator sequence. To achieve a strong expression a constitutive promoter construct was needed. Because of this Cauliflower Mosaik Virus [CMV] promotor was amplified by PCR using the BioBrick part BBa_J52038 (http://partsregistry.org/Part:BBa_J52038) and primers below-mentioned. The forward primer bound in the iGEM-prefix, while reverse primer bound at the end of the CMV-coding region. The reverse primer also contained an overhang to get the iGEM-suffix directly to the end of the promoter sequence which later allowed the cloning into the transfectionvector. CMV promoter was cloned into the vector by using the EcoRI and PstI restriction sites. To test the expression activity of the vector with CMV promoter the gene of the yellow fluorescent protein [YFP] was cloned behind the promoter region by using XbaI, PstI restriction enzymes for the YFP insert and SpeI, PstI for the vector to open Biobrick suffix. The tranfsfection of the resulting plasmid into 293T cells shows fully functionality.<br> |
<br><br> | <br><br> | ||
[[Image:Freiburg2008_TV_CMVRluc.jpg|700px]] | [[Image:Freiburg2008_TV_CMVRluc.jpg|700px]] | ||
Revision as of 10:23, 28 October 2008
 "
"