Team:UC Berkeley/Assembly
From 2008.igem.org
ChrisBrown (Talk | contribs) |
ChrisBrown (Talk | contribs) |
||
| Line 2: | Line 2: | ||
<div style="text-align: center;"><font size="6">'''Assembly'''</font></div><br> | <div style="text-align: center;"><font size="6">'''Assembly'''</font></div><br> | ||
| - | + | {{UCBmain}} | |
| + | <div style="text-align: center;"><font size="6">'''Assembly'''</font></div><br> | ||
| - | + | Our goal is to simplify two antibiotic assembly eliminating the replace mini-prep steps and making the protocol reagent-free. This will be accomplished by using our lysis device to lyse cells and extract DNA and engineering cells to produce their own restriction enzymes and ligase. | |
| - | + | For a general overview of two antibiotic assembly, click here [[https://2008.igem.org/Team:UC_Berkeley/LayeredAssembly]] | |
| - | + | ==''Assembly in Cell Lysate''== | |
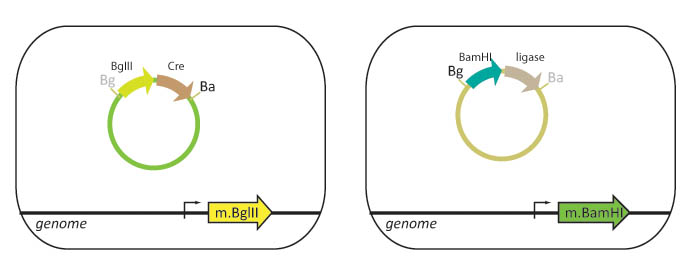
| - | + | Our lab currently using cells that are engineered to methylate either BamHI or BglII restriction sites. Part A is transformed into a "lefty" cell that is methylated on BglII restriction sites while Part B is transformed into a "righty" cell that is methylated on BamHI restriction sites. | |
| - | + | We propose to integrate the BamHI and BglII genes into the ''E. coli'' genome. In this scheme, lefty cells that are methylated on BglII cut sites, will stably express ligase and BglII. Righty cells that are methylated on BamHI cut sites, will be engineered to stably express XhoI and BamHI. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The methylation also protects the cellular DNA from being cut when these genes are expressed. | |
| - | In a single eppendorf tube, lefty and righty cells containing | + | In a single eppendorf tube, lefty and righty cells containing the plasmids described above are mixed and pelleted. The supernatant is discarded and the cells are resuspended in NEB2. The cells are then lysed using our lysis device. Click here for more information on our lysis device. [[https://2008.igem.org/Team:UC_Berkeley/LysisDevice]]. |
| + | |||
| + | The buffered lysate will contain all of the enzymes necessary for two antibiotic assembly. The lysate is incubated for one hour at 37 degrees and 30 minutes at 25 degrees. 10uL of lysate is transformed into new cells and the cells screened by plated on the proper antibiotic and growing overnight. | ||
| - | + | [[Image:cdb6.jpg]] | |
| - | + | We have begun testing of our BamHI and ligase strains of ''E. coli'' by using commercially available restriction enzymes and ligase. | |
| - | + | ==''Assembly in-Vivo''== | |
| + | |||
| + | We propose to engineer cells that are methylated on BamHI and BglII cut sites and stably express BamHI, BglII, Cre and ligase. These cells will be infected with two phagemids, each containing a basic part. | ||
Revision as of 01:48, 29 October 2008
Our goal is to simplify two antibiotic assembly eliminating the replace mini-prep steps and making the protocol reagent-free. This will be accomplished by using our lysis device to lyse cells and extract DNA and engineering cells to produce their own restriction enzymes and ligase.
For a general overview of two antibiotic assembly, click here [[1]]
Assembly in Cell Lysate
Our lab currently using cells that are engineered to methylate either BamHI or BglII restriction sites. Part A is transformed into a "lefty" cell that is methylated on BglII restriction sites while Part B is transformed into a "righty" cell that is methylated on BamHI restriction sites.
We propose to integrate the BamHI and BglII genes into the E. coli genome. In this scheme, lefty cells that are methylated on BglII cut sites, will stably express ligase and BglII. Righty cells that are methylated on BamHI cut sites, will be engineered to stably express XhoI and BamHI. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The methylation also protects the cellular DNA from being cut when these genes are expressed.
In a single eppendorf tube, lefty and righty cells containing the plasmids described above are mixed and pelleted. The supernatant is discarded and the cells are resuspended in NEB2. The cells are then lysed using our lysis device. Click here for more information on our lysis device. [[2]].
The buffered lysate will contain all of the enzymes necessary for two antibiotic assembly. The lysate is incubated for one hour at 37 degrees and 30 minutes at 25 degrees. 10uL of lysate is transformed into new cells and the cells screened by plated on the proper antibiotic and growing overnight.
We have begun testing of our BamHI and ligase strains of E. coli by using commercially available restriction enzymes and ligase.
Assembly in-Vivo
We propose to engineer cells that are methylated on BamHI and BglII cut sites and stably express BamHI, BglII, Cre and ligase. These cells will be infected with two phagemids, each containing a basic part.
 "
"