Team:Bologna/Software
From 2008.igem.org
(→Visual Fluo Bacteria: a software for the analysis of fluorescence bacteria image) |
(→A.R.Q.: an Automatic Registry Query generator software) |
||
| Line 103: | Line 103: | ||
= A.R.Q.: an Automatic Registry Query generator software= | = A.R.Q.: an Automatic Registry Query generator software= | ||
| - | The | + | The Registry is an important information source. It is aspected thet the amount of information will growth in the future, so we develop a java tool to search data in the Registry automatically. In facts the Registry page are well structured and write in a standard way, that enable automatic query by the simple reading of the html code. |
| - | + | The software has been developed in java language to ensure its indipendence from the machine type and from the OS.<br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | The software | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | The main form "SEARCH FORM" (figure 1) ha an user interface friendly and very simple: | ||
[[Image:Findpart.jpg|center|thumbnail|500px]] | [[Image:Findpart.jpg|center|thumbnail|500px]] | ||
<br><br> | <br><br> | ||
| + | By this form is possible to insert one of the following entry: | ||
| - | + | *"CODE PART":standard biobrick code (BBa_XXXXX) | |
| - | + | *"PARTIAL DESCRIPTION":any keyword that would be researched | |
| - | + | *"SEQUENCE":the sequence without space in one line | |
| - | the | + | By the drop-down menù "PART TYPE" it's possible to limit the search on: |
*measurement | *measurement | ||
*generator | *generator | ||
| Line 139: | Line 127: | ||
*terminator | *terminator | ||
*all<br> | *all<br> | ||
| - | + | ||
| - | + | Three query modalities are possible in this tool: | |
| - | When we use the | + | [[Image:tabellaingressorisultati.jpg|center|thumbnail|450px]] |
| - | + | <br><br> | |
| + | When we use the entry "PARTIAL DESCRIPTION" or "SEQUENCE" the result of the query could be not unique, so the result will show in the form "RESULT FORM" (figure 2): | ||
[[Image:finestra scelta.jpg|center|thumbnail|450px]] | [[Image:finestra scelta.jpg|center|thumbnail|450px]] | ||
<br> | <br> | ||
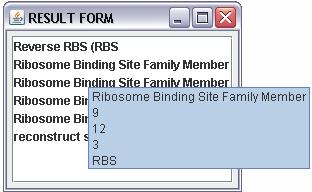
| - | + | "RESULT PAGE" shows a list of the registry parts that matched the entry, with a double click is possible to select the desired one; after this will be automatically upload in the "SEARCH FORM" the data part. | |
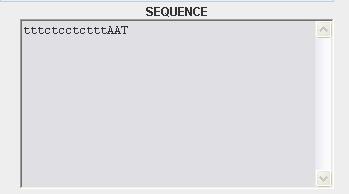
| - | possible | + | When the entry is "SEQUENCE" is possible to scroll the result in the "RESULT FORM" and will be visualized by a tool tip for every matched part (figure 3): |
| - | *the name of | + | |
| + | [[Image:tooltip.jpg|center|thumbnail|450px]] | ||
| + | |||
| + | <br><br> | ||
| + | *the name of part | ||
*the first and the last index of the alignment between the query and the answer sequence | *the first and the last index of the alignment between the query and the answer sequence | ||
*the length of the answer sequence | *the length of the answer sequence | ||
| - | *the group of the registry parts(regulatory,conjugation,signalling....) | + | *the group of the registry parts (regulatory,conjugation,signalling....) |
<br> | <br> | ||
| - | |||
| - | When a part is selected, by a double mouse double click, the program automatically update the | + | When a part is selected, by a double mouse double click, the program automatically update the data and change to upper case the shared nucleotide (figure 4). |
[[Image:sequencematch.jpg|center|thumbnail|450px]] | [[Image:sequencematch.jpg|center|thumbnail|450px]] | ||
Revision as of 09:35, 29 October 2008
| HOME | PROJECT | TEAM | SOFTWARE | MODELING | WET LAB | LAB-BOOK | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
Contents |
Visual Fluo Bacteria: a software for the analysis of fluorescence bacteria image
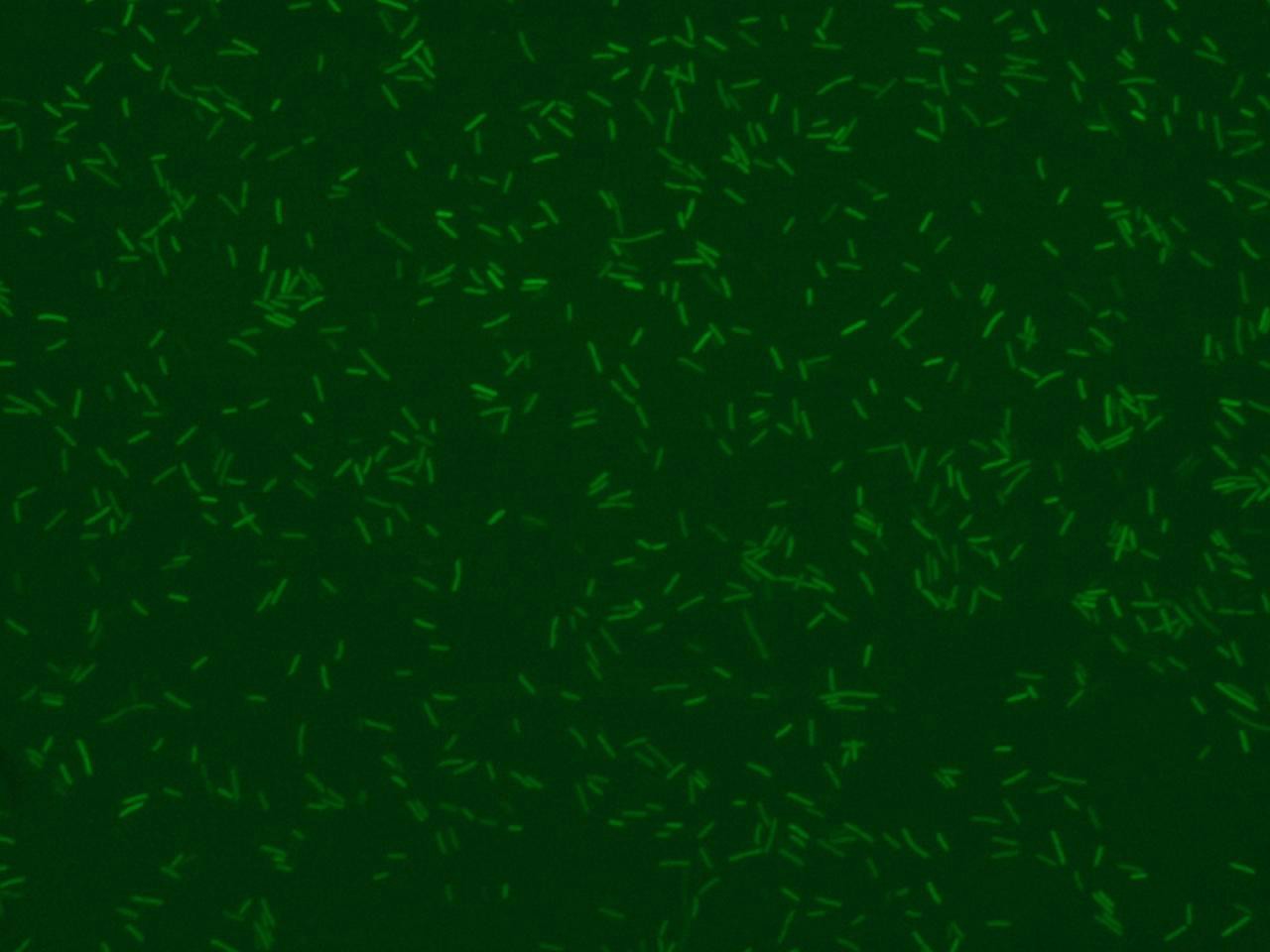
Promoter activity can be revealed using fluorescent proteins as reporters. An image of fluorescence bacteria can be obtain with a microscopy and a ccd camera ( Figure 1 shows an example of bacterial image).
The analysis of the image( scrivere il formato delle immagini) by the software allow us to achieve bacteria's morphology informations, size and the number of bacteria in a single view. Additionally we can assess the promoter activation dynamic in terms of fluorescence mean value for each bacterium and standard deviation.
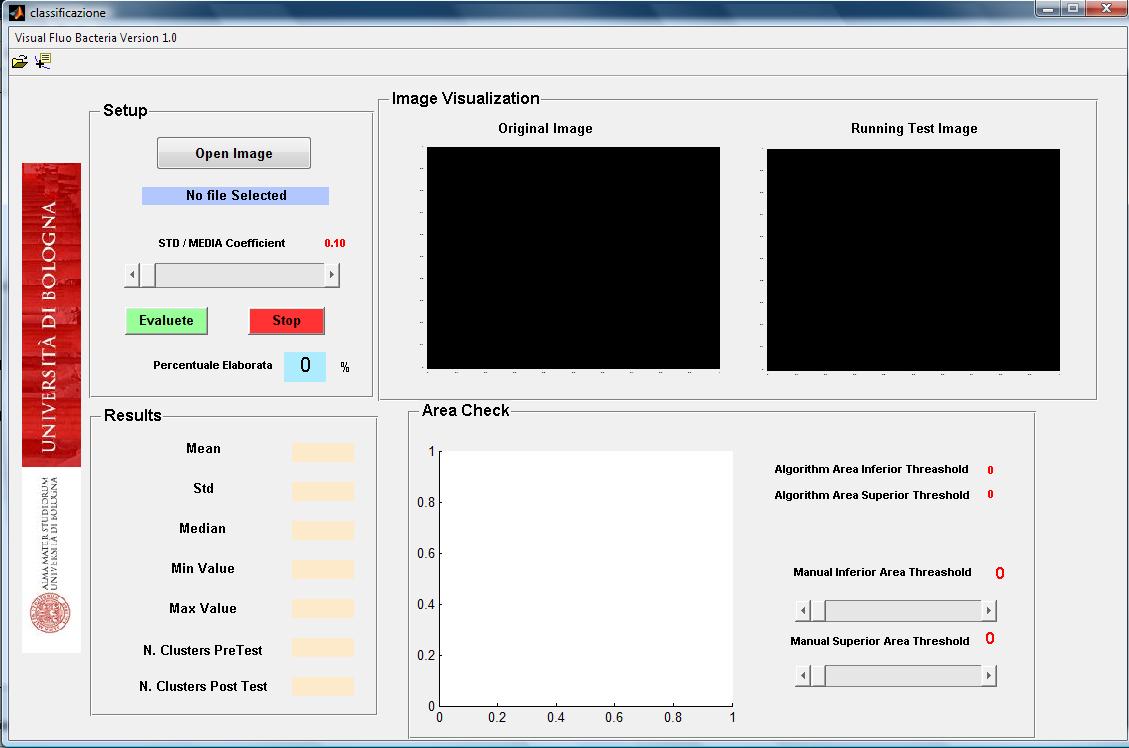
The user interface is shown in figure 2.
The use of the software is easy and intuitive.
The algorithm reads fluorescence images and converts it into a "black and white" one. Then the image is filtered by Top Hat filter to correct uneven illumination when the background is dark. In the next step the global threshold is computed in order to convert an intensity image to a binary image using Otsu’s method. The image is then ready to be scanned, pixel by pixel, to detect bacteria (cluster of pixels) and obtain informations about their area, fluorescence mean and standard deviation. Fluorescence is read from R channel for RFP, G for GFP and B for CFP. Then for each bacteria is computed size, mean and standard deviation.
Our software consists in a filtering of bacteria that are on the acquired image and it based on an algorithm that avail different parameters. In particular the user can set two indicators:
- Area dimension range ( definite by a superior and an inferior limit): This range defers to selected the bacteria that have similar size.
- Fluorescence intensity ( definite by the std\mean ratio referred to each bacteria): this values consent to throw away the bacteria that lie on another focal layer or that aren’t focused correctly.
At this point, the filter based on these values discard the bacteria that have a std\mean higher than the threeshold and the bacteria that have a dimension out of the range.
Example of filtering with very selective parameters (low ratio std/mean and narrow range of area dimensions)
|
All the obtained data are processed with area and focus efficiency parameters to estimate the population fluorescence mean, standard deviation, median, minimal and maximal fluorescence levels.
To download the program and relative user manual, you can click on the following icons.
![]() Visual Fluo Bacteria 1.0
Visual Fluo Bacteria 1.0
Note: after decompacting file, execute and run classificazione.m
![]() User Manual
User Manual
Microscopy system for fluorescence image acquisition
The system’s core is a Nikon Eclipse TE2000-U inverted fluorescence microscope. For GFP image acquisition we used a B-2A filter by Nikon with an excitation band between 450 and 490 nm and the optimal emission placed at 520 nm.
The illumination system is composed of a 75 Watt Xenon arc lamp connected to a Photon Technology Instruments DeltaRAM X monochromator. The control software( scrivere il nome) is implemented in a Labview environment and permits the regulation of the excitation wavelength and the calibration of the system.
The camera used to acquire images and film segments is a Nikon DS-5m with a DS-U1 controller. This one receives the acquired signal form the camera through a serial connection and sends it to the PC through an USB slot. Nikon also supplied an interface software for image acquisition and elaboration.
A.R.Q.: an Automatic Registry Query generator software
The Registry is an important information source. It is aspected thet the amount of information will growth in the future, so we develop a java tool to search data in the Registry automatically. In facts the Registry page are well structured and write in a standard way, that enable automatic query by the simple reading of the html code.
The software has been developed in java language to ensure its indipendence from the machine type and from the OS.
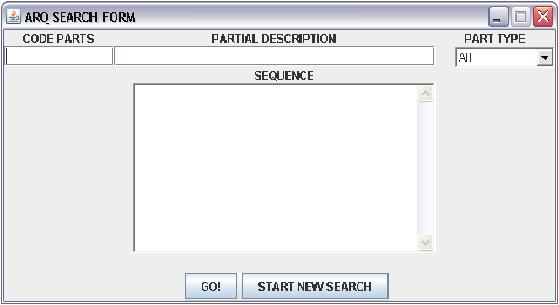
The main form "SEARCH FORM" (figure 1) ha an user interface friendly and very simple:
By this form is possible to insert one of the following entry:
- "CODE PART":standard biobrick code (BBa_XXXXX)
- "PARTIAL DESCRIPTION":any keyword that would be researched
- "SEQUENCE":the sequence without space in one line
By the drop-down menù "PART TYPE" it's possible to limit the search on:
- measurement
- generator
- composite
- rna
- dna
- conjugation
- reporters
- signalling
- rbs
- regulatory
- terminator
- all
Three query modalities are possible in this tool:
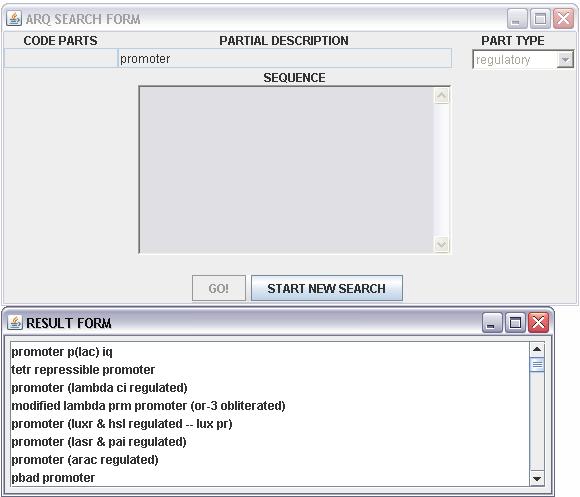
When we use the entry "PARTIAL DESCRIPTION" or "SEQUENCE" the result of the query could be not unique, so the result will show in the form "RESULT FORM" (figure 2):
"RESULT PAGE" shows a list of the registry parts that matched the entry, with a double click is possible to select the desired one; after this will be automatically upload in the "SEARCH FORM" the data part.
When the entry is "SEQUENCE" is possible to scroll the result in the "RESULT FORM" and will be visualized by a tool tip for every matched part (figure 3):
- the name of part
- the first and the last index of the alignment between the query and the answer sequence
- the length of the answer sequence
- the group of the registry parts (regulatory,conjugation,signalling....)
When a part is selected, by a double mouse double click, the program automatically update the data and change to upper case the shared nucleotide (figure 4).
 "
"