Team:KULeuven/Model/Filter
From 2008.igem.org
dock/undock dropdown
Contents |
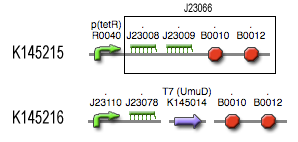
Filter
Position in the system
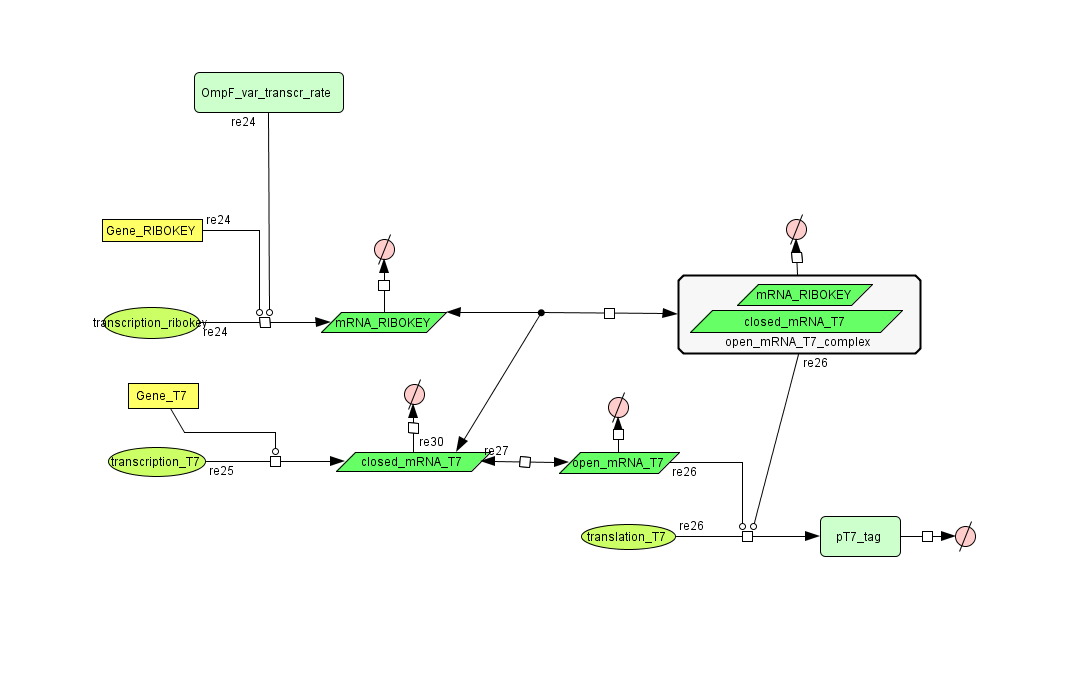
Describing the system
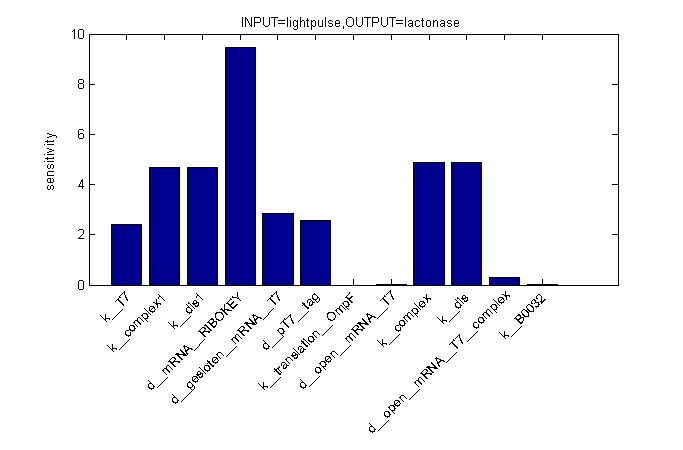
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation rates | |||
| dpT7_tag | 0.00155 s-1 | tag added to speed up degradation of otherwise too stable T7 polymerase | [http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP#Model_and_simulation link] |
| dmRNA_RIBOKEY | 0.00462 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dclosed_mRNA_T7 | 0.00462 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen_mRNA_T7 | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen_mRNA_T7_complex | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| Equilibrium constants | |||
| Keq_1 | 0,015 [M] | closed and open mRNA | |
| Keq_2 | 0.0212 [M] | closed and key (binded with ribokey) complex mRNA | |
| Transcription rates | |||
| kmRNA_RIBOKEY | Input dependent | ||
| kmRNA_T7 | 0,0011 s-1 | ||
Remark: The key-lock system can be enhanced to 0.3%-14% (todo: Jonas/Dries)
Models
CellDesigner (SBML file)
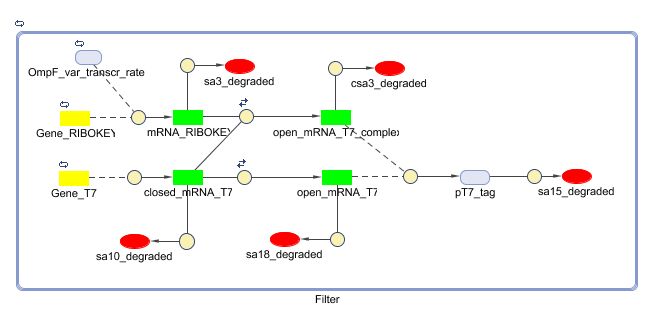
Matlab (SBML file)
Matlab diagram for the filter system:
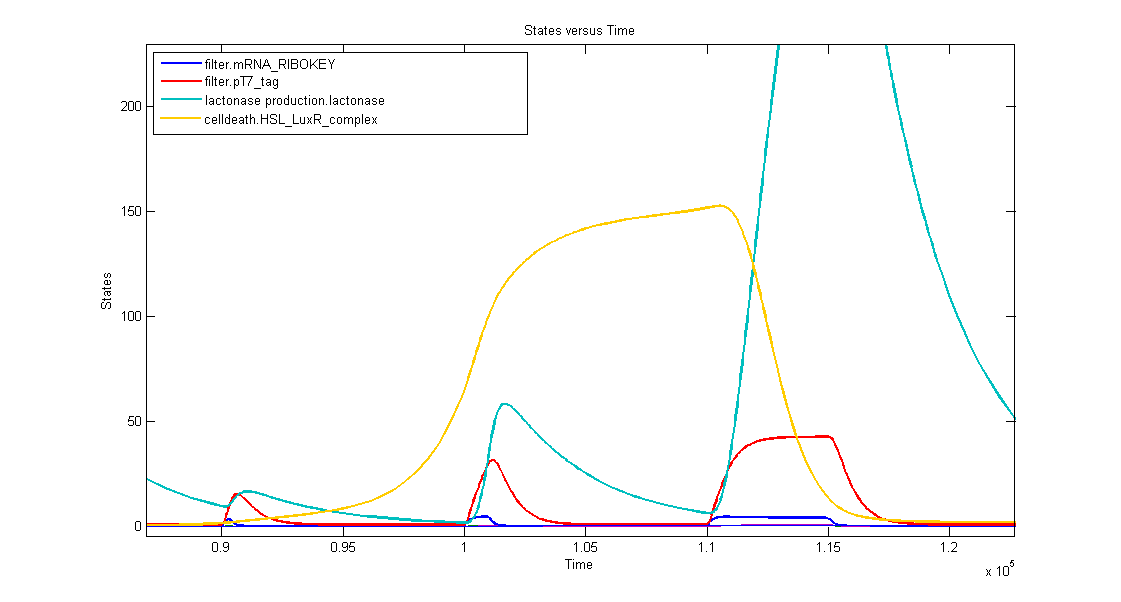
Simulations
 "
"