MODELING:
Reaction 4: Natural degradation of cI*
Half life of cI*:
The half-life of a modified cI is of 4 minutes, according to Elowitz & Leibler (2000). They analyze the LAA tail, and JB Andersen et al (1998) conclude that the LAA and LVA tails confer about the same time of life to GFP.
Reaction rate:
Once we get the half-life time of the protein, how do we calculate the rate of reaction and the flow?
The half-life of a reaction (t1/2) is the time it takes for half of the reagents to become products. In a first order reaction, t1/2 is a constant and can be calculated from the rate constant, as follows:
t1/2 =-ln(0.5)/k=0.693/k
This reciprocal relationship between the half life time and the rate constant is very useful when making an estimate of the time a reaction will take to occur. Thus, for k = 0.01/s, the half life time would be about 70 s. For k = 10/s, the half life would be of about 0.07 s or 70 milliseconds. The average life of the reactions of first order is also independent of the initial concentration. If the first half of the molecules react in aprox 20 s, half of the remaining molecules will also take 20 s to react, and so on. The fact that the average lifetime in an unimolecular reaction is a constant means that, at any time of the reaction, a constant fraction of reactive molecules have enough energy to overcome the kinetic barrier and become molecules of product. This makes sense because the energy of a set of molecules is distributed randomly according to a Boltzmann distribution.
* With information from RT Sauer (1999).
NOTE: A first order reaction is the type A → B.
Today we sent an e-mail to Dr. Peter Chivers, an expert in RcnR, to ask him for information in regards to RcnA.
Our mail to Dr. Peter Chivers:
Dr. Peter Chivers,
Hi! My name is Carlos and I'm a student of the Undergraduate Program in Genomic Sciences of the National Autonomous University of Mexico and currently I'm on the fourth year.
The main reason of this e-mail is to get in contact with you, I'll explain you why as briefly as I can. Several students of the program decided to participate in a systems biology competition known as iGEM, and our project focuses on regulating nickel efflux in e. coli. We have to both build the circuit and design a mathematical model to accurately predict the behaviour of our system.
Taking several issues in consideration we decided to regulate RcnA.
Here is the page of our team in case you're interested in reading more about our project:
https://2008.igem.org/Team:LCG-UNAM-Mexico
However we've been having some trouble in finding parameters for our model (such as RcnA's half-life, efficiency, synthesis rate among others). Therefore, having exhausted (or so we believe) the literary resources and having found your name in many of the papers regarding RcnA we decided to contact you. I'll try to explain with more detail the parameters we still lack in case you're interested in providing us with information.
I greatly thank you for taking the time to read this e-mail and await your answer.
Carlos Vargas
Dr. Peter Chivers' reply
Dear Carlos,
Thank you for your e-mail. From what I read on your website, it looks like your team is tackling an interesting project. Unfortunately, there are very few papers on RcnA itself, and none of them have examined the parameters that you need for your model. As far as I know, the groups that have published in the RcnR/RcnA area don't typically do those sorts of measurements so they are unlikely that have the data you need. We have no plans to do these experiments as my lab focuses on the nickel responsive regulators, RcnR and NikR.
I'm sorry that I can't be of more assistance. Good luck with your project, I hope it is well received at the iGEM competition.
Best regards,
Peter
WET LAB
Agarose Gel 1% low fusion point for band purification
- Molecular Weight Marker
- [1] Cage2 /11_1 pBB+RcnA
- [2] Cage1 /1_2 pBB+RcnA
- [5] Cage2b /8_1 pBB+RcnA
- [9] Cage2b /4_1 pBB+RcnA
Gel Purification PCR of pBB+rcnA
- Purification 1
- Purification 2
- Purification 3
- Purification 4 clean PRK
- Positive control
- Negative control
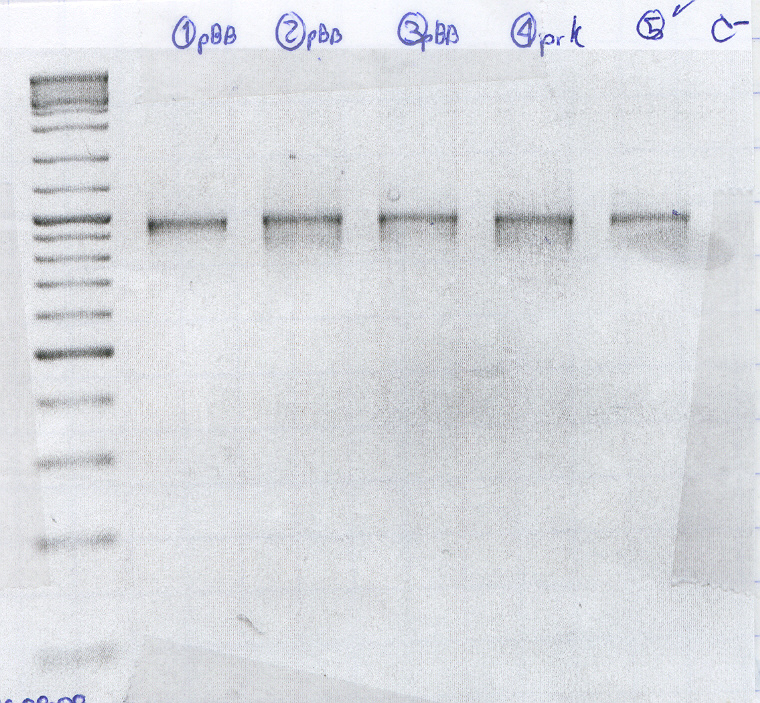
The cells seem to carry the plasmid with RcnA, However one of the negative controls gave product.
All of them were positive, however there are some spurious products.(no photo was added because the low melting gels are quite fragile and we couldn't carry it to the transiluminator with camera.)
Recipe:
DNA |
2 μl |
Water |
32 μl |
dNTP’s |
2.5 μl |
Mg(Cl)2 |
2.5 μl |
Oligo 1up |
2.5 μl |
Oligo 2low |
2.5 μl |
Taq |
1 μl |
Buffer |
5 μl |
A gel with the extraction of the plasmid PRK415+part_1 was run.
Gel Electrophoresis of PCR_Ligation part1+part2
- [5] 1_2 oligo 1up-2lower
- [4] 1_2 oligo 1up-2lower
- [3] 1_2 oligo 1up-2lower
- Molecular marker
- [2] 1_2 oligo 1up-2lower
- [1] 1_2 oligo 1up-2lower

We have got part1+part2 ligation



