Team:Bologna
From 2008.igem.org
| HOME | PROJECT | TEAM | SOFTWARE | MODELING | WET LAB | LAB-BOOK | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
| contenuto |

| 
|
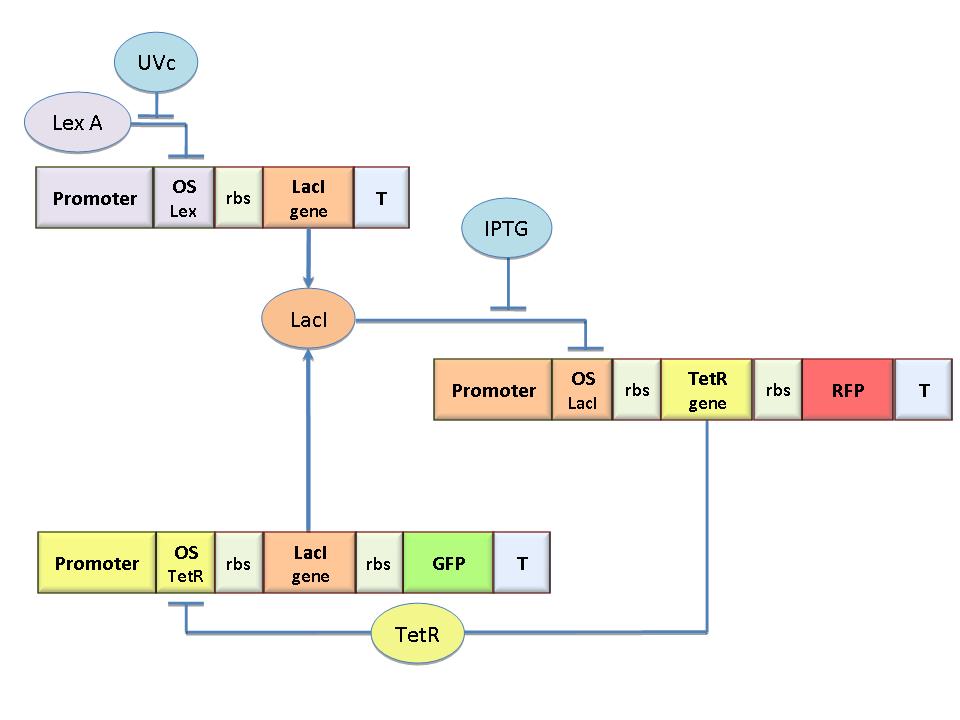
Our project aims to design a bacterial reprogrammable memory with genetically engineered E.coli colonies in solid medium working as an array of binary memory cells. To engineer bacteria we designed a genetic flip-flop (SR Latch) composed of a binary memory and an UV sensitive trigger. We chose UV to have a fine spatial selectivity in programming the cells and IPTG to reset all the memory cells. We designed the circuit by model-based analysis. Core elements of the genetic memory are two mutually regulated promoters. Each promoter has as an operator site flanking a constitutive promoter. Thus, promoter transcriptional strength and repressor binding affinity can be independently fixed. Operators for LacI, TetR, Lambda and LexA repressors were cloned as BioBricks to allow the rational design of regulated promoters that is still lacking in the Registry. A simple procedure was established to characterize the regulated promoter. We expect these parts to be a benefit in many Synthetic Biology applications. Click here for more information...
The rationale underpinning the fundations of the Ecoli.PROM team consists in the synergistical collaboration of a number of various young experts in precise areas of science. Such elements will feed into the group the expertise and know-how in Biotechnology, Electronic and Biomedical engineering with the additional support of pharmacists and models experts.
The innovation will indeed lie in this compound effort, directed towards the driving of synthetic biology to implement a system working as a result of complex biological activity, rather than conventional electronic elaboration. Click here for more information...
The aim of this work area is to determine the characteristics of the promoters that form the circuit. In this section are also collected a brief description of our model, all the equations that describe analytically our circuit, the most meaningful graphics and the results of simulations. Enjoy yourself!
Into this section you can find the very useful softwares we developed for this competition. The first of them realizes a fluorescence image analysis using two parameters: the bacteria dimensions and the standard deviation and mean fluorescence ratio. The second makes possible to find the registered parts you are searching for in a quick and easy way, to identify their sequence and display parts with similar behaviour. You can also download them from this page.
Here's all our lab work: week by week you can find all the adopted procedures, links to the registry of standard parts and protocols. The chronological structure of this section, organized as a notebook, mirrors the real development of our project and respects the pure iGEM style.
Go to the page!
Aknowledgements
Our Team is funded by:
- [http://www.unibo.it/Portale/default.htm University of Bologna]
- [http://serinar.criad.unibo.it Ser.In.Ar. Cesena]
 "
"