Team:ESBS-Strasbourg/Project idea history
From 2008.igem.org
Brainstormings
Before choosing to construct the cell cycle dependent toggle switch for this years iGEM competition we had many brainstorming sessions. Following is a list and short description of some of the ideas we had. If you want interested in one of the ideas, go ahead and construct.
E-Language
The E-language is an approach to minimize the interaction between a given system and a host organism. The complete transcription and translational machinery from E.Coli will be expressed in yeast. Then an already existing system from the Registry designed for the use in prokaryotic organism can be used in yeast, without adapting e.g. activation domains and repressors to the eukaryotic expression system.
Biological Milling Machine (BMM)
The goal of this idea is to construct a system for E.coli, so that they can serve as "intelligent" material for constructing structures in 3D space.
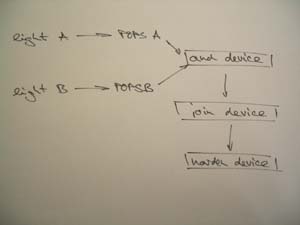
Therefore, two different light sources are directed from two different angles on E.coli containing glassware. Absorption of only both light sources will induce a program in bacteria to join with neighbour bacteria and secondly to harden (see figure BMM Devices). Consequently, a solid connected structure can be obtained.
In terms of the genetic program four modules are foreseen. Another challenge will be presented by requirements of the matrix.
Random Device
The goal is to construct a device which allows a single bacteria or a colony to respond to starting signal with several equiprobable output signals whereby the choice for one output is imposing all other outputs.
Throwing a dice, presuming an ideal player, monitors three main characteristics. First, the result is random, that means totally independent from the environment respectively the player. Secondly, all six outcomes have the exact probability to occur and third, one outcome is excluding the other five. One of our projects ideas for the iGEM 08 competition was transferring the properties of throwing a dice into a biological system. This, we foresaw to do in a strict sense of synthetic biology and the Biobrick standards. Hence, an distinct box, including all necessary biological parts would be constructed which offers the possibility of the attachment of other Biobricks which can serve as input or use the resulting outcome.
In order to keep the complexity of the system to a minimum our goal would have been to construct a device, which is showing two equipropable, mutually exclusive outputs in response to an input, the starting signal. Difficult might have been to find two molecular components which trigger further events with the same probability and this also under the changing conditions. Therefore, we thought about using twice the same component which has in turn different subsequent events as the transcription of green or red fluorescent protein. The random device is not only a biological alternative to an ordinary dice but we think that it is also representing an important brick for synthetic biology, since decisions are done without any interference from outside.
More in detail we were thinking about two promoters of the lexA type coupled to red respectively green fluorescent protein. In order that the transcription is only occurring of one protein we will nest the two promoter sequences into each other so that binding of one TF is hindering binding of a TF to the second promoter. Further one has to keep in mind that the binding of a TF is relatively short lasting a jumping to the second promoter is likely. In order to increase the binding time of the TF to the promoter we wanted to link the TF to a second molecule with DNA binding properties, thus representing a two feet molecule.
Single Serving Detector
Several strains are planned to be constructed whereby each of them has the property to detect a certain toxin or pesticide. In this respect the detector will work like an inverse enterotube.
Magnetotaxis
Magneto tactic bacteria have developed in many different classes of independent bacteria. They all have together that they built up a organelle in which they stock magnetic iron. Therefore they will align them always with the earth magnetic field. Based on these biological exceptions, an organelle in procaryotes and the use of magnetic fields, we thought about several possibilities:
- transferring those organelles in E. coli. This could have served to purify proteins by targeting them to the organelle membrane and afterwards separate them with the help of the magnetic properties of the organelle.
- another idea was to use the alignment of magneto tactic bacteria directly. This could have resulted in a compass, or a sort of hard disk ...
- a last idea was to grow up those bacteria and gain the build up small magnets, as they are used directly in many applications in the industry and research.
 "
"