Team:Freiburg/3D-Modeling
From 2008.igem.org
(Difference between revisions)
| Line 5: | Line 5: | ||
<br><br> | <br><br> | ||
== DNA-Origami == | == DNA-Origami == | ||
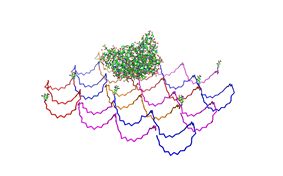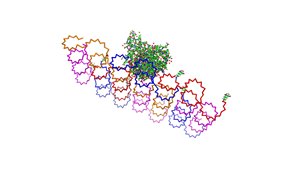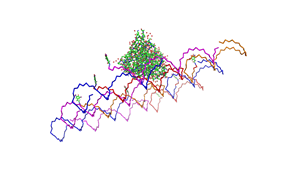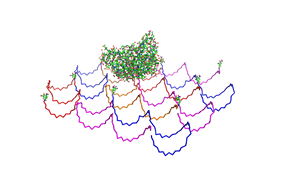
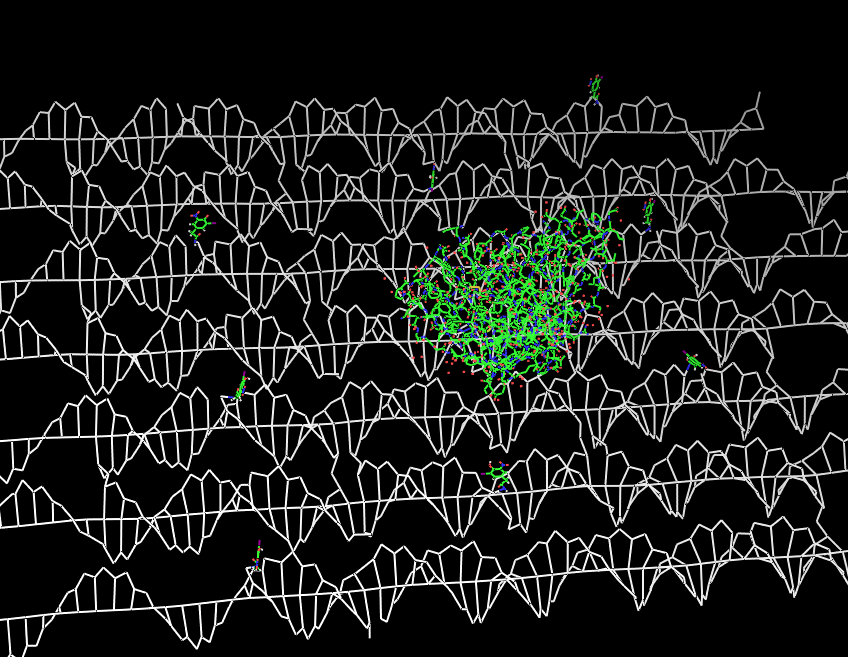
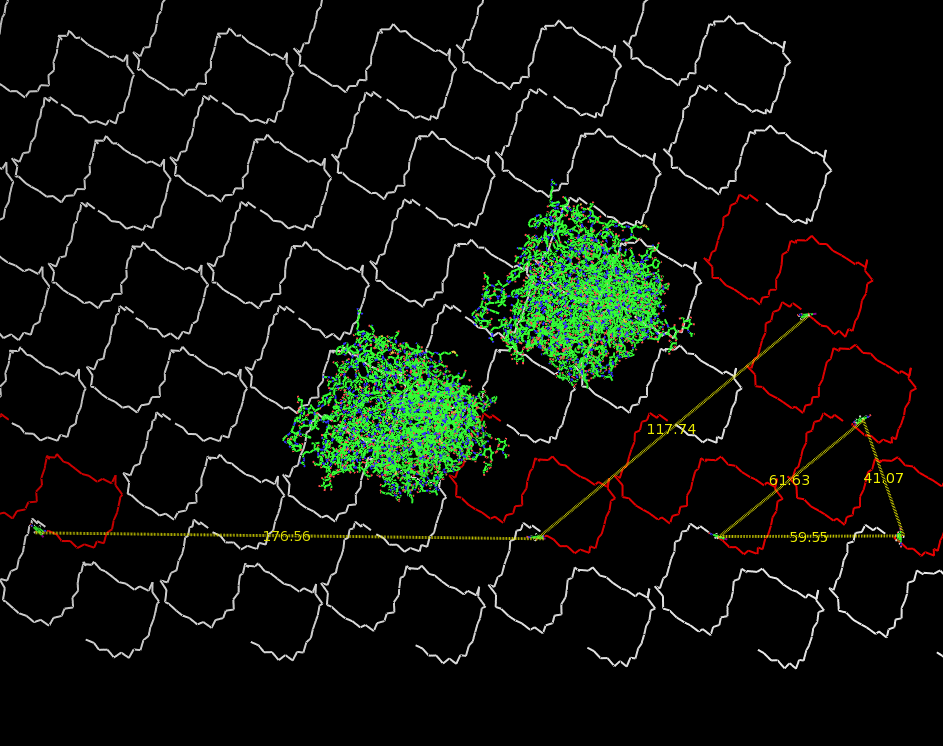
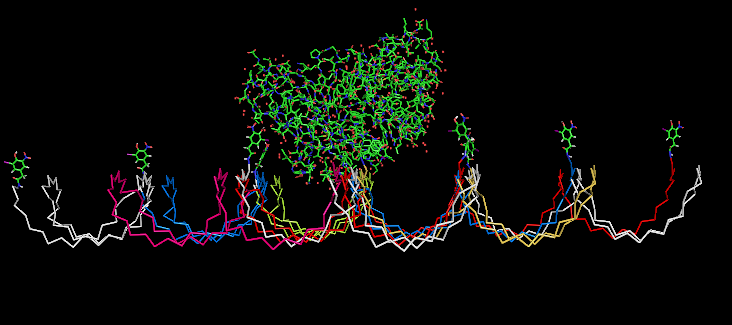
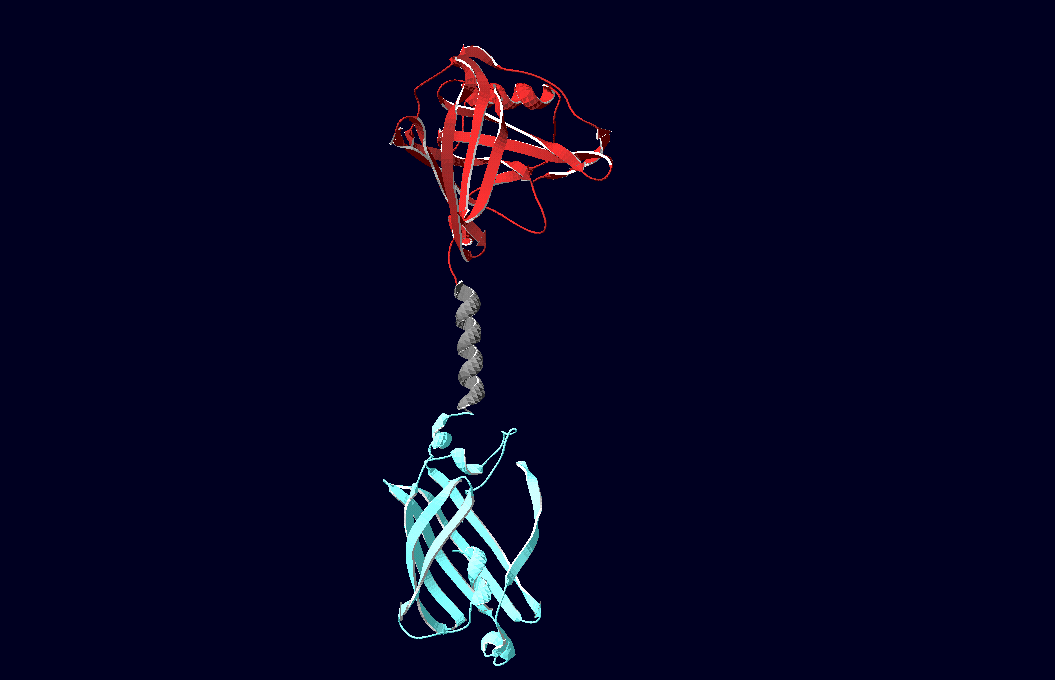
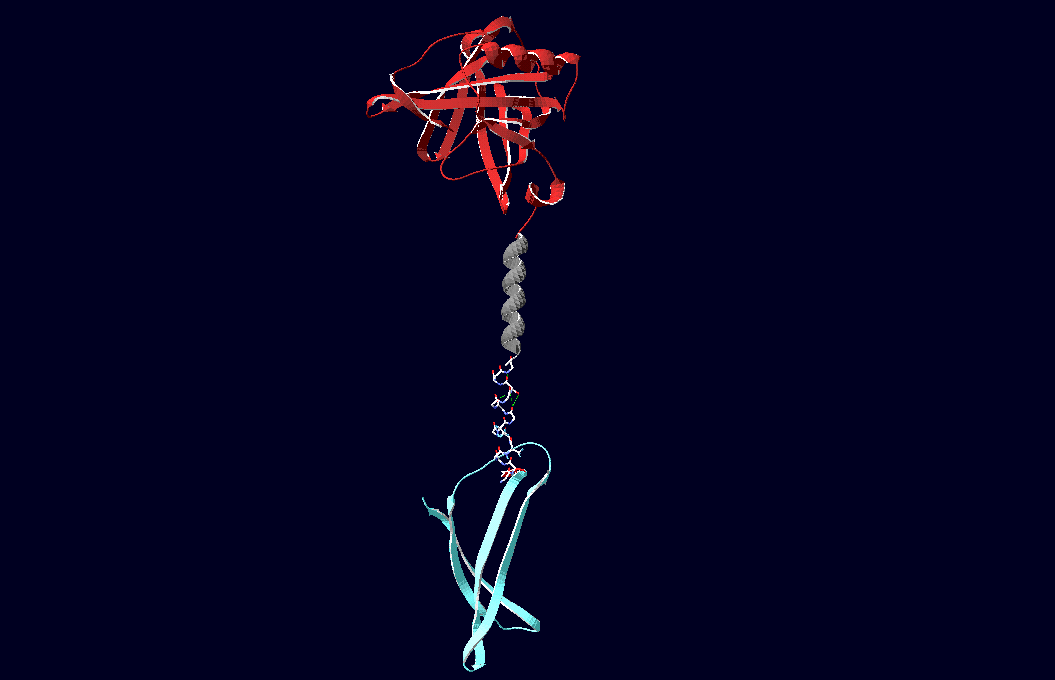
| - | Planning the input pattern on the DNA-Origami surface, we generated various 3D-models of the huge molecule using "Nano- | + | Planning the input pattern on the DNA-Origami surface, we generated various 3D-models of the huge molecule using "Nano-Engineer" (a free program in its beta-version), Pymol and SwissPdbViewer (both freeware). Some of those models are shown here to give you an impression of the molecules we have engineered this year.<br> |
[[Image:Freiburg2008_Fab_on_Origami_animated.gif|800 px]] | [[Image:Freiburg2008_Fab_on_Origami_animated.gif|800 px]] | ||
'''Fig.1:'''Some of the oligo nucleotides that shape the single-strand template fused to NIP-molecules and an anti-NIP-Fab-Fragment<br><br><br> | '''Fig.1:'''Some of the oligo nucleotides that shape the single-strand template fused to NIP-molecules and an anti-NIP-Fab-Fragment<br><br><br> | ||
Revision as of 22:34, 28 October 2008
 "
"