Team:KULeuven/11 September 2008
From 2008.igem.org
(→Modeling) |
m |
||
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| + | |||
| + | {{:Team:KULeuven/Tools/New_Day/Date_Retriever}} | ||
== Lab Work == | == Lab Work == | ||
| Line 24: | Line 27: | ||
Finalising and rounding up. Still messing with the sensitivity analysis... | Finalising and rounding up. Still messing with the sensitivity analysis... | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 01:46, 30 October 2008
| << return to notebook | return to homepage >> | ||
| < previous friday | ← yesterday | tomorrow → | next monday > |
Contents |
Lab Work
Wet Lab
We electroporated the following parts: K145201+(R0040+P0353), C0062+(K145150+B0034), K145253+K145254, (I712074+J23032)+(E0040+B0015), (B0014+B0033)+(K145151+B0015), (J23116+B0032)+(C0062+B0015), (J23109+J23032)+(K145001+B0015) and B0015+K145013.
We made a new ligation mix: J23116+B0034.
We made some digests with SpeI: J23109 and I712074.
The oligos to make the new lock arrived today. So we started the end-filling procedure to make this part (J23078) using Klenow polymerase.
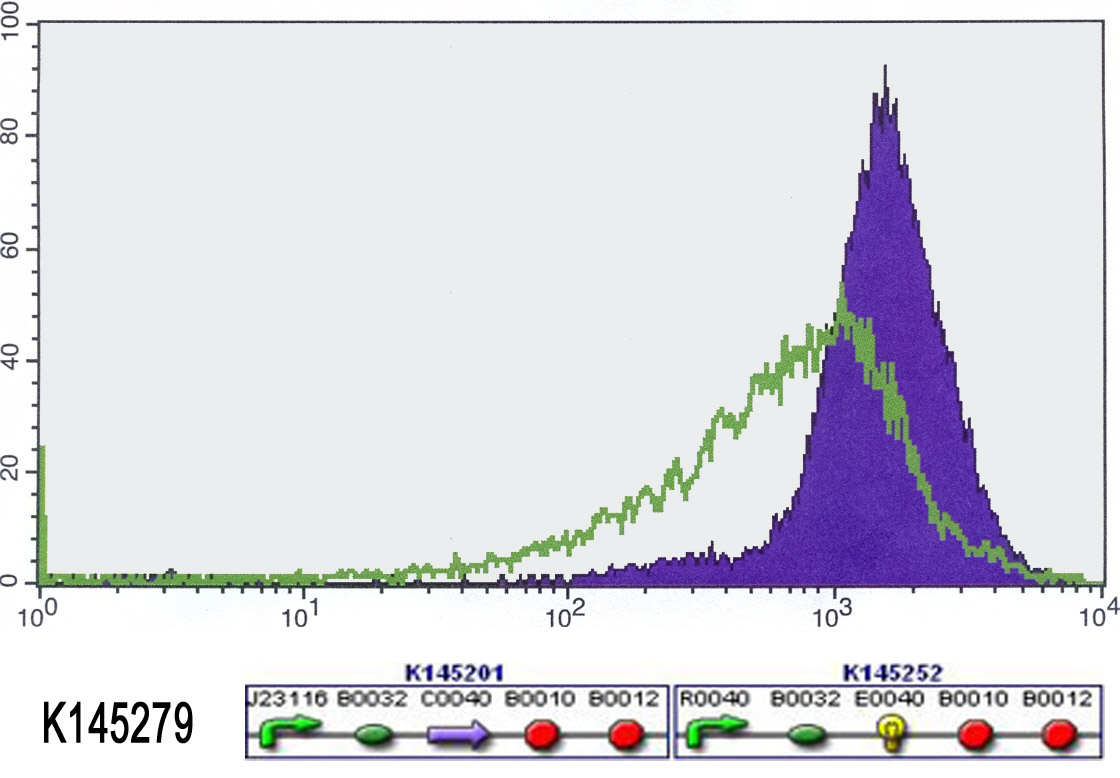
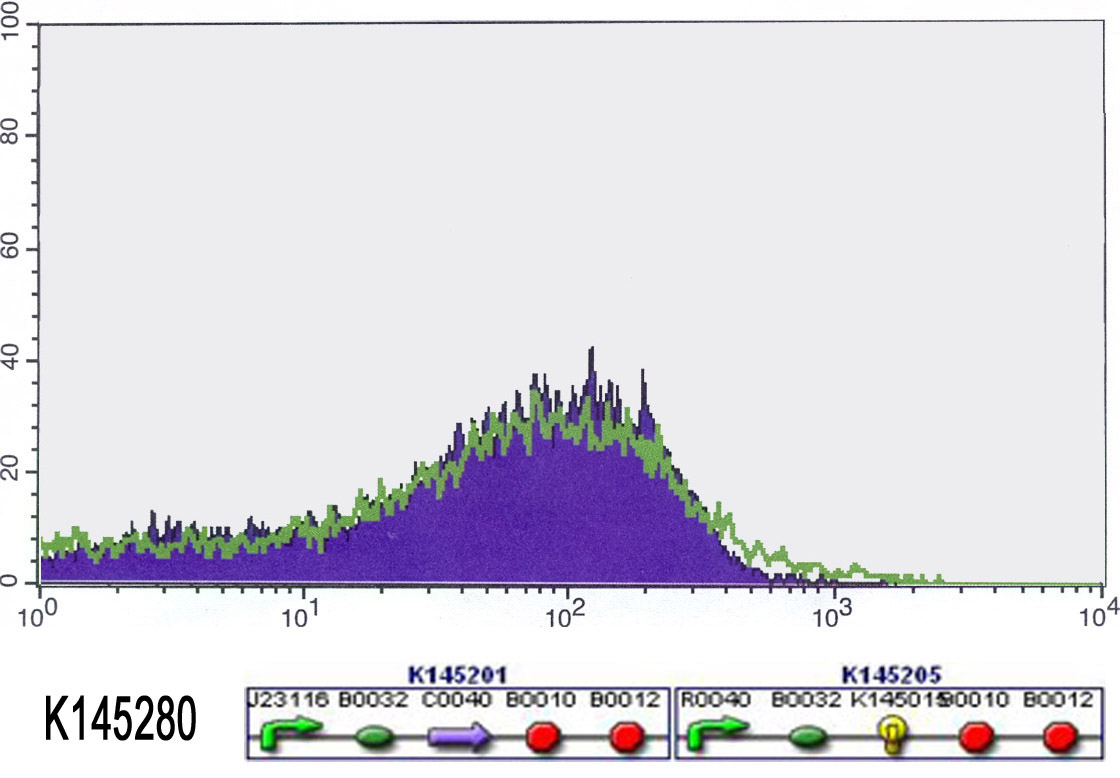
We also started to measure our input-output systems (K145279 and K145280) using FACS. Normally, when no anhydrotetracyclin is present, the cells shouldn't fluoresce. Unfortunately, the did already fluoresce from the start. We added anhydrotetracyclin (50 ng/ml) to the liquid cultures and allowed the cells to grow in this aTc medium for 4 hours. Then we measured the fluorescence again. Ideally, adding aTc would switch on the fluorescence. So we expected that there would be more fluorescence. This was not the case, we measured just as much fluorescence as in the beginning. The results can be found in the figures below. Because the cells always fluoresce, the outputs definitely work. But we can not switch them on or off. This means that there is something wrong with the input. Maybe it wasn't properly ligated (-> sequencing)? Maybe there is not enough repressor (-> stronger RBS)? Maybe the repressor doesn't work (-> big problem)?
Dry Lab
Modeling
Finalising and rounding up. Still messing with the sensitivity analysis...
 "
"