Team:KULeuven/8 August 2008
From 2008.igem.org
(gels) |
m |
||
| Line 13: | Line 13: | ||
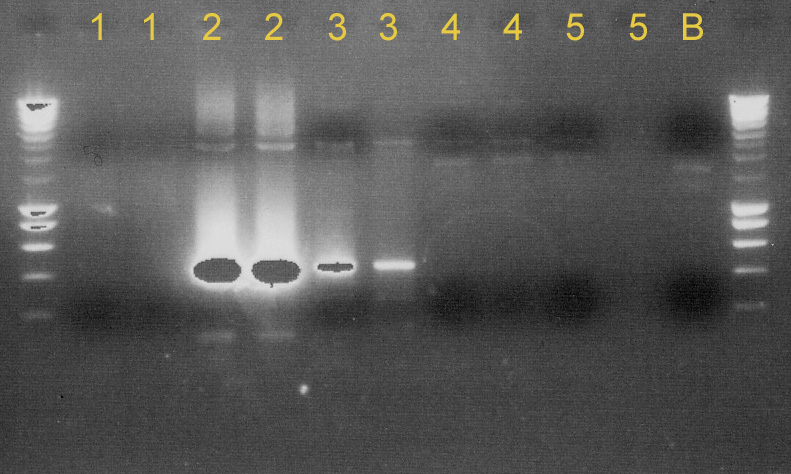
We tested [http://partsregistry.org/Part:BBa_C0040 C0040]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_P1010 P1010]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_C0060 C0060]+[http://partsregistry.org/Part:BBa_B0015 B0015] and [http://partsregistry.org/Part:BBa_J23100 J23100]+[http://partsregistry.org/Part:BBa_B0032 B0032] to see if the ligation succeeded. We used ''taq'' polymerase and primers VF/VR-2. The results were disappointing. None of them were properly ligated. (see '''Gel 1'''). | We tested [http://partsregistry.org/Part:BBa_C0040 C0040]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_P1010 P1010]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_C0060 C0060]+[http://partsregistry.org/Part:BBa_B0015 B0015] and [http://partsregistry.org/Part:BBa_J23100 J23100]+[http://partsregistry.org/Part:BBa_B0032 B0032] to see if the ligation succeeded. We used ''taq'' polymerase and primers VF/VR-2. The results were disappointing. None of them were properly ligated. (see '''Gel 1'''). | ||
| - | We made minipreps of some other ligated parts ([http://partsregistry.org/Part:BBa_E0022 E0022]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_J23109 J23109]+[http://partsregistry.org/Part:BBa_J23032 J23032], [http://partsregistry.org/Part:BBa_I712074 I712074]+[http://partsregistry.org/Part:BBa_J23032 J23032], [http://partsregistry.org/Part:BBa_K145015 K145015]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_K145015 K145015]+[http://partsregistry.org/Part: | + | We made minipreps of some other ligated parts ([http://partsregistry.org/Part:BBa_E0022 E0022]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_J23109 J23109]+[http://partsregistry.org/Part:BBa_J23032 J23032], [http://partsregistry.org/Part:BBa_I712074 I712074]+[http://partsregistry.org/Part:BBa_J23032 J23032], [http://partsregistry.org/Part:BBa_K145015 K145015]+[http://partsregistry.org/Part:BBa_B0015 B0015], [http://partsregistry.org/Part:BBa_K145015 K145015]+[http://partsregistry.org/Part:pSB1A2 pSB1A2] and [http://partsregistry.org/Part:BBa_C0062 C0062]+[http://partsregistry.org/Part:BBa_B0015 B0015]). After that, we did a PCR using the VR/VF2 primers and ''taq'' polymerase to see whether these ligations worked or not. E0022+B0015, K145015+pSB1A2 and C0062+B0015 were OK. J23109+J23032, I712074+J23032 and K145015+B0015 gave mixed results. Some of the colonies contained ligated parts, others did not. So we have to try to isolate the right colonies. (see '''Gel 2'''). |
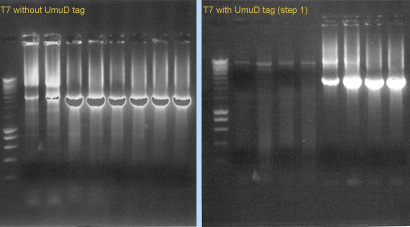
We also started to construct the T7 RNA polymerase (with and whithout UmuD tag), using PCR. We used ''pfx'' polymerase and the primers 1076/1077 (for T7 without tag) or 1078/1077 (for T7 with tag). The template was part [http://partsregistry.org/Part:BBa_I712022 I712022]. (see '''Gel 3'''). | We also started to construct the T7 RNA polymerase (with and whithout UmuD tag), using PCR. We used ''pfx'' polymerase and the primers 1076/1077 (for T7 without tag) or 1078/1077 (for T7 with tag). The template was part [http://partsregistry.org/Part:BBa_I712022 I712022]. (see '''Gel 3'''). | ||
Latest revision as of 19:53, 10 October 2008
| << return to notebook | return to homepage >> | ||
| < previous friday | ← yesterday | tomorrow → | next monday > |
Contents |
Lab Work
Wet Lab
We tested C0040+B0015, P1010+B0015, C0060+B0015 and J23100+B0032 to see if the ligation succeeded. We used taq polymerase and primers VF/VR-2. The results were disappointing. None of them were properly ligated. (see Gel 1).
We made minipreps of some other ligated parts (E0022+B0015, J23109+J23032, I712074+J23032, K145015+B0015, K145015+pSB1A2 and C0062+B0015). After that, we did a PCR using the VR/VF2 primers and taq polymerase to see whether these ligations worked or not. E0022+B0015, K145015+pSB1A2 and C0062+B0015 were OK. J23109+J23032, I712074+J23032 and K145015+B0015 gave mixed results. Some of the colonies contained ligated parts, others did not. So we have to try to isolate the right colonies. (see Gel 2).
We also started to construct the T7 RNA polymerase (with and whithout UmuD tag), using PCR. We used pfx polymerase and the primers 1076/1077 (for T7 without tag) or 1078/1077 (for T7 with tag). The template was part I712022. (see Gel 3).
We electroporated some ligated parts: R0084+B0032, R0062+B0032, R0084+J23022 and C0012+B0015.
We made a liquid culture of parts B0032, C0060, C0062, P1010 and R0040. On Monday, we can miniprep them. We also made a liquid culture of Top10 cells to make new electrocompetent cells.
Dry Lab
General
Hybrid promoter in front of ccdB is not enough because the timer will have started. So the safest way might be to use antisense LuxI (leaky though it is) and the hybrid promoter. If it's too much work/costs, just antisense LuxI maybe.
The following parts will have to be on a higher copy number plasmid (preferably a pSB5X#): antisense LuxI, the ribokey and the 434 without the LVA tag.
Modeling
Worked some more on the sensitivity analyses with a bunch of new strange results... Probably have to think of a better way to represent sensitivity, the formula of ETH Zürich 2007 may not apply that good to our situation. Apart from this, we're still waiting for the first lab-results to come in so we can verify how representative the parameters in the simulation are.
 "
"