Team:KULeuven/Model/Inverter
From 2008.igem.org
(→Parameters) |
m (→Describing the system) |
||
| (25 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
| + | {{:Team:KULeuven/Tools/Scripting}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| - | + | [[Image:pictogram_inverter.png|120px|right]] | |
| + | |||
==Invertimer== | ==Invertimer== | ||
=== Position in the system === | === Position in the system === | ||
| - | The | + | The InverTimer subsystem receives its input from the filter, T7. The InverTimer's function is to '''produce''' HSL when '''no''' input is present, so a low T7 input gives rise to a high HSL output and vice versa. The production of HSL means that the cell will start a timer that eventually will be used in the celldeath-subsystem to produce ccdB. In this way the cell will die off if no desease remains present. |
=== Describing the system === | === Describing the system === | ||
| - | see also: [https://2008.igem.org/Team:KULeuven/Project/Inverter Project: | + | see also: [https://2008.igem.org/Team:KULeuven/Project/Inverter Project:InverTimer] |
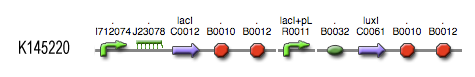
[[Image:Inverter_BioBrick.jpg|center]] | [[Image:Inverter_BioBrick.jpg|center]] | ||
| Line 18: | Line 21: | ||
<body> | <body> | ||
<p> | <p> | ||
| - | <a href="https://static.igem.org/mediawiki/2008/ | + | <a href="https://static.igem.org/mediawiki/2008/3/35/InverterODE.pdf"> |
<img border="0" src="https://2008.igem.org/wiki/skins/common/images/icons/fileicon-pdf.png" width="65" height="60"> | <img border="0" src="https://2008.igem.org/wiki/skins/common/images/icons/fileicon-pdf.png" width="65" height="60"> | ||
</a> | </a> | ||
| Line 39: | Line 42: | ||
| d<sub>LVA</sub> = 2.814E-4 s<sup>-1</sup> | | d<sub>LVA</sub> = 2.814E-4 s<sup>-1</sup> | ||
| LVA-tag reduces lifetime to 40 minutes | | LVA-tag reduces lifetime to 40 minutes | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html> [7<html>]</html>] |
|- | |- | ||
| d<sub>RNA_LuxI</sub> | | d<sub>RNA_LuxI</sub> | ||
| 0.0025 s<sup>-1</sup> | | 0.0025 s<sup>-1</sup> | ||
| | | | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [5<html>]</html>] |
|- | |- | ||
| d<sub>LuxI_antimRNA</sub> | | d<sub>LuxI_antimRNA</sub> | ||
| 0.0045303737 s<sup>-1</sup> | | 0.0045303737 s<sup>-1</sup> | ||
| estimate: because this RNA isn't translated, it degrades faster | | estimate: because this RNA isn't translated, it degrades faster | ||
| - | | | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [5<html>]</html>] |
|- | |- | ||
| d<sub>LacI</sub> | | d<sub>LacI</sub> | ||
| d<sub>LVA</sub> = 2.814E-4 s<sup>-1</sup> | | d<sub>LVA</sub> = 2.814E-4 s<sup>-1</sup> | ||
| LVA-tag reduces lifetime to 40 minutes | | LVA-tag reduces lifetime to 40 minutes | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html> [7<html>]</html>] |
|- | |- | ||
| d<sub>closed mRNA LacI</sub> | | d<sub>closed mRNA LacI</sub> | ||
| 0.0046209812 s<sup>-1</sup> | | 0.0046209812 s<sup>-1</sup> | ||
| estimate: because this mRNA isn't translated, it degrades faster | | estimate: because this mRNA isn't translated, it degrades faster | ||
| - | | | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [5<html>]</html>] |
|- | |- | ||
| d<sub>open mRNA LacI</sub> | | d<sub>open mRNA LacI</sub> | ||
| 0.0023104906 s<sup>-1</sup> | | 0.0023104906 s<sup>-1</sup> | ||
| | | | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [5<html>]</html>] |
|- | |- | ||
| d<sub>open mRNA LacI complex</sub> | | d<sub>open mRNA LacI complex</sub> | ||
| 0.0023104906 s<sup>-1</sup> | | 0.0023104906 s<sup>-1</sup> | ||
| | | | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [5<html>]</html>] |
|- | |- | ||
| d<sub>HSL</sub> | | d<sub>HSL</sub> | ||
| 1.02E-6 s<sup>-1</sup> | | 1.02E-6 s<sup>-1</sup> | ||
| very stable in the medium, lifetime around 185h | | very stable in the medium, lifetime around 185h | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [11<html>]</html>] |
| + | |- | ||
| + | ! colspan="4" style="border-bottom: 1px solid #003E81;" | LuxI catalysis | ||
| + | |- | ||
| + | | k<sub>cat</sub> | ||
| + | | 0.0166666667 s<sup>-1</sup> | ||
| + | | Estimated to be about 90% of V<sub>max</sub> in LB medium. | ||
| + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [4<html>]</html>] | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | T7 Transcription | ! colspan="4" style="border-bottom: 1px solid #003E81;" | T7 Transcription | ||
| Line 81: | Line 91: | ||
| 421 | | 421 | ||
| dissociation constant, recalculated to remove units | | dissociation constant, recalculated to remove units | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [10<html>]</html>] |
|- | |- | ||
| k<sub>max</sub> | | k<sub>max</sub> | ||
| 0.044 s<sup>-1</sup> | | 0.044 s<sup>-1</sup> | ||
| maximal T7 transcription rate | | maximal T7 transcription rate | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [10<html>]</html>] |
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Key-Lock constants | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Key-Lock constants | ||
| Line 92: | Line 102: | ||
| K<sub>eq 1</sub> | | K<sub>eq 1</sub> | ||
| 0,015 [M] | | 0,015 [M] | ||
| - | | between closed and open T7 mRNA, experimental | + | | between closed and open T7 mRNA, modeled for competition, experimental |
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
|- | |- | ||
| K<sub>eq 2</sub> | | K<sub>eq 2</sub> | ||
| 0.0212 [M] | | 0.0212 [M] | ||
| - | | between closed T7 mRNA and key unlocked mRNA complex, experimental | + | | between closed T7 mRNA and key unlocked mRNA complex, modeled for competition, experimental |
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
|- | |- | ||
| k<sub>dis2</sub> | | k<sub>dis2</sub> | ||
| - | | | + | | 0.00416 s<sup>-1</sup> |
| derived from experimental values | | derived from experimental values | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
|- | |- | ||
| k<sub>complex2</sub> | | k<sub>complex2</sub> | ||
| - | | | + | | 0.00237 s<sup>-1</sup> |
| derived from experimental values | | derived from experimental values | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
|- | |- | ||
| k<sub>closed</sub> | | k<sub>closed</sub> | ||
| - | | | + | | 500 s<sup>-1</sup> |
| derived from experimental values | | derived from experimental values | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
|- | |- | ||
| k<sub>open</sub> | | k<sub>open</sub> | ||
| - | | | + | | 7.5 s<sup>-1</sup> |
| derived from experimental values | | derived from experimental values | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | LacI repression | ! colspan="4" style="border-bottom: 1px solid #003E81;" | LacI repression | ||
| Line 130: | Line 135: | ||
| 1.0E-10 M<sup>-1</sup> | | 1.0E-10 M<sup>-1</sup> | ||
| Dissociation constant | | Dissociation constant | ||
| - | | | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [3<html>]</html>] |
|- | |- | ||
| n<sub>LacI</sub> | | n<sub>LacI</sub> | ||
| 2.0 | | 2.0 | ||
| Hill coefficient for LacI | | Hill coefficient for LacI | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [3<html>]</html>] |
|- | |- | ||
| k_trans_LacI | | k_trans_LacI | ||
| - | | 0. | + | | 0.0025 s<sup>-1</sup> |
| Estimated maximal transcription rate from R0011 | | Estimated maximal transcription rate from R0011 | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [9<html>]</html>] |
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Antisense LuxI | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Antisense LuxI | ||
|- | |- | ||
| k_complex3 | | k_complex3 | ||
| - | | | + | | 0.00237 s<sup>-1</sup> |
| - | | rate constant for formation of asRNA - LuxI mRNA duplex | + | | rate constant for formation of asRNA - LuxI mRNA duplex |
| - | | | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [3<html>]</html>] |
|- | |- | ||
| K<sub>mRNA_LuxI:antisense_mRNA</sub> | | K<sub>mRNA_LuxI:antisense_mRNA</sub> | ||
| 4.22E14 | | 4.22E14 | ||
| Complex of LuxI mRNA with antisense mRNA | | Complex of LuxI mRNA with antisense mRNA | ||
| - | | | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [1<html>]</html>] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
! colspan="4" style="border-bottom: 1px solid #003E81;" | Translation Rates | ! colspan="4" style="border-bottom: 1px solid #003E81;" | Translation Rates | ||
| Line 164: | Line 164: | ||
| 0.167 s<sup>-1</sup> | | 0.167 s<sup>-1</sup> | ||
| translation rate for B0032 RBS (0.3 relative efficiency) | | translation rate for B0032 RBS (0.3 relative efficiency) | ||
| - | | [ | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [8<html>]</html>] |
|- | |- | ||
| k<sub>transl LacI</sub> | | k<sub>transl LacI</sub> | ||
| 0.167 s<sup>-1</sup> | | 0.167 s<sup>-1</sup> | ||
| - | | lock defined translation rate for LacI | + | | estimate: lock defined translation rate for LacI |
| - | | | + | | [https://2008.igem.org/Team:KULeuven/Model/Inverter#References [2<html>]</html>] |
|} | |} | ||
| Line 179: | Line 179: | ||
==== Matlab ([https://static.igem.org/mediawiki/2008/0/00/Inverter_Matlab.zip SBML file])==== | ==== Matlab ([https://static.igem.org/mediawiki/2008/0/00/Inverter_Matlab.zip SBML file])==== | ||
| - | + | [[Image:Inverter_Matlab.jpg|700px|center|Inverter]] | |
| - | [[Image:Inverter_Matlab.jpg| | + | |
=== Simulations === | === Simulations === | ||
| - | |||
| - | + | {| class="wikitable" | |
| + | |- | ||
| + | !width="80"| Time span | ||
| + | !width="80"| Input (TetR) | ||
| + | !width="720"| Results | ||
| + | |- | ||
| + | | A | ||
| + | | 0.0125 s<sup>-1</sup> | ||
| + | | The amount LacI increases from state zero to state one because both mRNA_RIBOKEY and pT7_tag are present. This results in a repression of LuxI which decreases to zero: the input signal (TetR) is inverted. | ||
| + | |- | ||
| + | | B | ||
| + | | 5E-5 s<sup>-1</sup> | ||
| + | | The amount LacI decreases back to state zero. The amount LuxI remains the same (state zero). | ||
| + | |- | ||
| + | | C | ||
| + | | 5E-5 s<sup>-1</sup> | ||
| + | | LuxI changes from state zero to state one. Time span B and C form together the transient behaviour of the inverter when the input signal changes from one to zero. | ||
| + | |- | ||
| + | | D | ||
| + | | 5E-5 s<sup>-1</sup> | ||
| + | | LuxI remains in state one: the input signal is once again inverted. | ||
| + | |- | ||
| + | | E | ||
| + | | 0.0125 s<sup>-1</sup> | ||
| + | | A short pulse of 1000 seconds has a influence a steep decrease of LuxI. | ||
| + | |- | ||
| + | | F & G | ||
| + | | 5E-5 s<sup>-1</sup> | ||
| + | | During time span F and G, LuxI decreases further for a while and increases back to state one. | ||
| + | |- | ||
| + | | H | ||
| + | | 5E-5 s<sup>-1</sup> | ||
| + | | LuxI is back in state one. | ||
| + | |} | ||
| - | + | The simulation shows a working inverter (left figure). A small disadvantage is the transient behaviour of the inverter: a small pulse of 1000 seconds results in a transient behaviour of +- 30000 seconds. Also for a long pulse (10000 seconds) is a long transient behaviour noticeable (40000 seconds). The effect of the inverter on the timer aspect is visuable in the right figure: a long pulse ( from 10000 till 11000) resets the timer (HSL drecreases till zero). After this pulse and the transient behaviour of the inverter, the timer restarts counting. The short pulse (from 200000 till 201000 seconds) only partially resets the timer. | |
| - | [[ | + | All graphs have amounts (number of molecules in the cell) plotted vs time, measured in seconds. |
| + | |||
| + | <html> | ||
| + | <div class="center"> | ||
| + | <div class="noborder" style="overflow: auto; width: 800px; height: 520px;"> | ||
| + | <div class="noborder" style="width: 1200px;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/d/d1/Sim_inverter_1.png" style="float: left; width: 550px; height: 500px; margin: 0 5px;" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/2/27/HSL.png" style="float: left; width: 600px; height: 500px; margin: 0 5px;" /> | ||
| + | </div></div></div></html> | ||
| + | |||
| + | === References === | ||
| + | |||
| + | <html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en"> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8"/> | ||
| + | <title>Bibliography</title> | ||
| + | </head> | ||
| + | <body> | ||
| + | <table style="border-collapse:collapse;line-height:1.1em;"> | ||
| + | <tr style="vertical-align:top;"><td>[1]</td><td style="padding-left:4pt;">A. E G H Wagner and R W Simons, “Antisense RNA Control in Bacteria, Phages, and Plasmids,” Nov. 2003; http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.mi.48.100194.003433.</td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[2]</td><td style="padding-left:4pt;">“Berkeley2006-RiboregulatorsMain - IGEM”; http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain.</td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | |||
| + | <tr style="vertical-align:top;"><td>[3]</td><td style="padding-left:4pt;">“ETHZ/Parameters - IGEM07”; https://2007.igem.org/ETHZ/Parameters.</td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[4]</td><td style="padding-left:4pt;">“Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein,” Sep. 1996; http://www.pnas.org/content/93/18/9505.</td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[5]</td><td style="padding-left:4pt;">J.A. Bernstein et al., “Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays,” <span style="font-style:italic;">Proceedings of the National Academy of Sciences of the United States of America</span>, vol. 99, Jul. 2002, pp. 9697–9702. <span class="Z3988" title="url_ver=Z39.88-2004&ctx_ver=Z39.88-2004&rft_id=info%3Adoi/10.1073/pnas.112318199&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Global%20analysis%20of%20mRNA%20decay%20and%20abundance%20in%20Escherichia%20coli%20at%20single-gene%20resolution%20using%20two-color%20fluorescent%20DNA%20microarrays&rft.jtitle=Proceedings%20of%20the%20National%20Academy%20of%20Sciences%20of%20the%20United%20States%20of%20America&rft.stitle=Proc%20Natl%20Acad%20Sci%20U%20S%20A.%20&rft.volume=99&rft.issue=15&rft.aufirst=Jonathan%20A.&rft.aulast=Bernstein&rft.au=Jonathan%20A.%20Bernstein&rft.au=Arkady%20B.%20Khodursky&rft.au=Pei-Hsun%20Lin&rft.au=Sue%20Lin-Chao&rft.au=Stanley%20N.%20Cohen&rft.date=2002-07-23&rft.pages=9697%E2%80%939702"></span></td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[6]</td><td style="padding-left:4pt;">W. Hsieh et al., “Influence of sequence and distance between two operators on interaction with the lac repressor,” <span style="font-style:italic;">J. Biol. Chem.</span>, vol. 262, Oct. 1987, pp. 14583-14591. <span class="Z3988" title="url_ver=Z39.88-2004&ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Influence%20of%20sequence%20and%20distance%20between%20two%20operators%20on%20interaction%20with%20the%20lac%20repressor&rft.jtitle=J.%20Biol.%20Chem.&rft.volume=262&rft.issue=30&rft.aufirst=WT&rft.aulast=Hsieh&rft.au=WT%20Hsieh&rft.au=PA%20Whitson&rft.au=KS%20Matthews&rft.au=RD%20Wells&rft.date=1987-10-25&rft.pages=14583-14591"></span></td></tr> | ||
| + | |||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[7]</td><td style="padding-left:4pt;">J.B. Andersen et al., “New Unstable Variants of Green Fluorescent Protein for Studies of Transient Gene Expression in Bacteria,” <span style="font-style:italic;">Applied and Environmental Microbiology</span>, vol. 64, Jun. 1998, pp. 2240–2246. <span class="Z3988" title="url_ver=Z39.88-2004&ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=New%20Unstable%20Variants%20of%20Green%20Fluorescent%20Protein%20for%20Studies%20of%20Transient%20Gene%20Expression%20in%20Bacteria&rft.jtitle=Applied%20and%20Environmental%20Microbiology&rft.stitle=Appl%20Environ%20Microbiol.%20&rft.volume=64&rft.issue=6&rft.aufirst=Jens%20Bo&rft.aulast=Andersen&rft.au=Jens%20Bo%20Andersen&rft.au=Claus%20Sternberg&rft.au=Lars%20Kongsbak%20Poulsen&rft.au=Sara%20Petersen%20Bj%C3%B8rn&rft.au=Michael%20Givskov&rft.au=S%C3%B8ren%20Molin&rft.date=1998-06&rft.pages=2240%E2%80%932246"></span></td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[8]</td><td style="padding-left:4pt;">“Part:BBa B0032 - partsregistry.org”; http://partsregistry.org/Part:BBa_B0032.</td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[9]</td><td style="padding-left:4pt;">“Part:BBa R0011 - partsregistry.org”; http://partsregistry.org/Part:BBa_R0011.</td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[10]</td><td style="padding-left:4pt;">G.M. Skinner et al., “Promoter Binding, Initiation, and Elongation By Bacteriophage T7 RNA Polymerase: A SINGLE-MOLECULE VIEW OF THE TRANSCRIPTION CYCLE,” <span style="font-style:italic;">J. Biol. Chem.</span>, vol. 279, Jan. 2004, pp. 3239-3244. <span class="Z3988" title="url_ver=Z39.88-2004&ctx_ver=Z39.88-2004&rft_id=info%3Adoi/10.1074/jbc.M310471200&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Promoter%20Binding%2C%20Initiation%2C%20and%20Elongation%20By%20Bacteriophage%20T7%20RNA%20Polymerase%3A%20A%20SINGLE-MOLECULE%20VIEW%20OF%20THE%20TRANSCRIPTION%20CYCLE&rft.jtitle=J.%20Biol.%20Chem.&rft.volume=279&rft.issue=5&rft.aufirst=Gary%20M.&rft.aulast=Skinner&rft.au=Gary%20M.%20Skinner&rft.au=Christoph%20G.%20Baumann&rft.au=Diana%20M.%20Quinn&rft.au=Justin%20E.%20Molloy&rft.au=James%20G.%20Hoggett&rft.date=2004&rft.pages=3239-3244"></span></td></tr> | ||
| + | |||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | <tr style="vertical-align:top;"><td>[11]</td><td style="padding-left:4pt;">Y. Wang and J.R. Leadbetter, “Rapid Acyl-Homoserine Lactone Quorum Signal Biodegradation in Diverse Soils,” <span style="font-style:italic;">Appl. Environ. Microbiol.</span>, vol. 71, Mar. 2005, pp. 1291-1299. <span class="Z3988" title="url_ver=Z39.88-2004&ctx_ver=Z39.88-2004&rft_id=info%3Adoi/10.1128/AEM.71.3.1291-1299.2005&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Rapid%20Acyl-Homoserine%20Lactone%20Quorum%20Signal%20Biodegradation%20in%20Diverse%20Soils&rft.jtitle=Appl.%20Environ.%20Microbiol.&rft.volume=71&rft.issue=3&rft.aufirst=Ya-Juan&rft.aulast=Wang&rft.au=Ya-Juan%20Wang&rft.au=Jared%20Renton%20Leadbetter&rft.date=2005-03-01&rft.pages=1291-1299"></span></td></tr> | ||
| + | <tr><td colspan="2"> </td></tr> | ||
| + | </table></body> | ||
| + | </html> | ||
Latest revision as of 23:20, 29 October 2008
Contents |
Invertimer
Position in the system
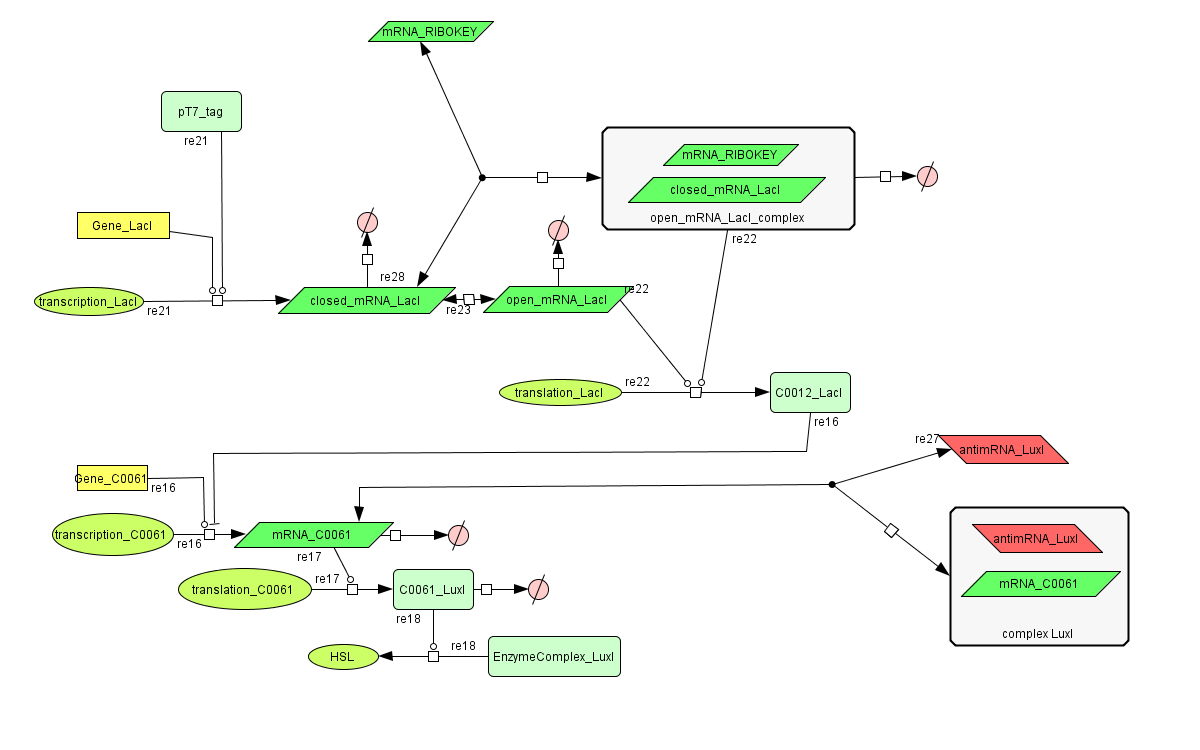
The InverTimer subsystem receives its input from the filter, T7. The InverTimer's function is to produce HSL when no input is present, so a low T7 input gives rise to a high HSL output and vice versa. The production of HSL means that the cell will start a timer that eventually will be used in the celldeath-subsystem to produce ccdB. In this way the cell will die off if no desease remains present.
Describing the system
see also: Project:InverTimer
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dLuxI | dLVA = 2.814E-4 s-1 | LVA-tag reduces lifetime to 40 minutes | [2] [7] |
| dRNA_LuxI | 0.0025 s-1 | [5] | |
| dLuxI_antimRNA | 0.0045303737 s-1 | estimate: because this RNA isn't translated, it degrades faster | [5] |
| dLacI | dLVA = 2.814E-4 s-1 | LVA-tag reduces lifetime to 40 minutes | [2] [7] |
| dclosed mRNA LacI | 0.0046209812 s-1 | estimate: because this mRNA isn't translated, it degrades faster | [5] |
| dopen mRNA LacI | 0.0023104906 s-1 | [5] | |
| dopen mRNA LacI complex | 0.0023104906 s-1 | [5] | |
| dHSL | 1.02E-6 s-1 | very stable in the medium, lifetime around 185h | [11] |
| LuxI catalysis | |||
| kcat | 0.0166666667 s-1 | Estimated to be about 90% of Vmax in LB medium. | [4] |
| T7 Transcription | |||
| KT7 | 421 | dissociation constant, recalculated to remove units | [10] |
| kmax | 0.044 s-1 | maximal T7 transcription rate | [10] |
| Key-Lock constants | |||
| Keq 1 | 0,015 [M] | between closed and open T7 mRNA, modeled for competition, experimental | [2] |
| Keq 2 | 0.0212 [M] | between closed T7 mRNA and key unlocked mRNA complex, modeled for competition, experimental | [2] |
| kdis2 | 0.00416 s-1 | derived from experimental values | [2] |
| kcomplex2 | 0.00237 s-1 | derived from experimental values | [2] |
| kclosed | 500 s-1 | derived from experimental values | [2] |
| kopen | 7.5 s-1 | derived from experimental values | [2] |
| LacI repression | |||
| KLacI | 1.0E-10 M-1 | Dissociation constant | [3] |
| nLacI | 2.0 | Hill coefficient for LacI | [3] |
| k_trans_LacI | 0.0025 s-1 | Estimated maximal transcription rate from R0011 | [9] |
| Antisense LuxI | |||
| k_complex3 | 0.00237 s-1 | rate constant for formation of asRNA - LuxI mRNA duplex | [3] |
| KmRNA_LuxI:antisense_mRNA | 4.22E14 | Complex of LuxI mRNA with antisense mRNA | [1] |
| Translation Rates | |||
| ktransl LuxI | 0.167 s-1 | translation rate for B0032 RBS (0.3 relative efficiency) | [8] |
| ktransl LacI | 0.167 s-1 | estimate: lock defined translation rate for LacI | [2] |
Models
CellDesigner (SBML file)
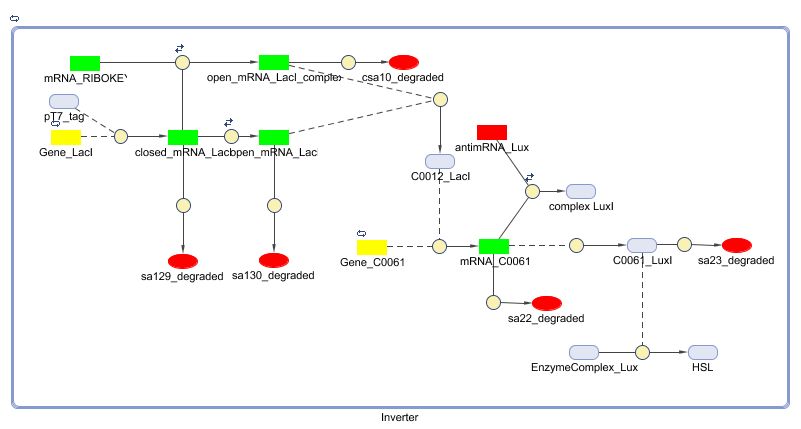
Matlab (SBML file)
Simulations
| Time span | Input (TetR) | Results |
|---|---|---|
| A | 0.0125 s-1 | The amount LacI increases from state zero to state one because both mRNA_RIBOKEY and pT7_tag are present. This results in a repression of LuxI which decreases to zero: the input signal (TetR) is inverted. |
| B | 5E-5 s-1 | The amount LacI decreases back to state zero. The amount LuxI remains the same (state zero). |
| C | 5E-5 s-1 | LuxI changes from state zero to state one. Time span B and C form together the transient behaviour of the inverter when the input signal changes from one to zero. |
| D | 5E-5 s-1 | LuxI remains in state one: the input signal is once again inverted. |
| E | 0.0125 s-1 | A short pulse of 1000 seconds has a influence a steep decrease of LuxI. |
| F & G | 5E-5 s-1 | During time span F and G, LuxI decreases further for a while and increases back to state one. |
| H | 5E-5 s-1 | LuxI is back in state one. |
The simulation shows a working inverter (left figure). A small disadvantage is the transient behaviour of the inverter: a small pulse of 1000 seconds results in a transient behaviour of +- 30000 seconds. Also for a long pulse (10000 seconds) is a long transient behaviour noticeable (40000 seconds). The effect of the inverter on the timer aspect is visuable in the right figure: a long pulse ( from 10000 till 11000) resets the timer (HSL drecreases till zero). After this pulse and the transient behaviour of the inverter, the timer restarts counting. The short pulse (from 200000 till 201000 seconds) only partially resets the timer.
All graphs have amounts (number of molecules in the cell) plotted vs time, measured in seconds.


References
| [1] | A. E G H Wagner and R W Simons, “Antisense RNA Control in Bacteria, Phages, and Plasmids,” Nov. 2003; http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.mi.48.100194.003433. |
| [2] | “Berkeley2006-RiboregulatorsMain - IGEM”; http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain. |
| [3] | “ETHZ/Parameters - IGEM07”; https://2007.igem.org/ETHZ/Parameters. |
| [4] | “Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein,” Sep. 1996; http://www.pnas.org/content/93/18/9505. |
| [5] | J.A. Bernstein et al., “Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays,” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, Jul. 2002, pp. 9697–9702. |
| [6] | W. Hsieh et al., “Influence of sequence and distance between two operators on interaction with the lac repressor,” J. Biol. Chem., vol. 262, Oct. 1987, pp. 14583-14591. |
| [7] | J.B. Andersen et al., “New Unstable Variants of Green Fluorescent Protein for Studies of Transient Gene Expression in Bacteria,” Applied and Environmental Microbiology, vol. 64, Jun. 1998, pp. 2240–2246. |
| [8] | “Part:BBa B0032 - partsregistry.org”; http://partsregistry.org/Part:BBa_B0032. |
| [9] | “Part:BBa R0011 - partsregistry.org”; http://partsregistry.org/Part:BBa_R0011. |
| [10] | G.M. Skinner et al., “Promoter Binding, Initiation, and Elongation By Bacteriophage T7 RNA Polymerase: A SINGLE-MOLECULE VIEW OF THE TRANSCRIPTION CYCLE,” J. Biol. Chem., vol. 279, Jan. 2004, pp. 3239-3244. |
| [11] | Y. Wang and J.R. Leadbetter, “Rapid Acyl-Homoserine Lactone Quorum Signal Biodegradation in Diverse Soils,” Appl. Environ. Microbiol., vol. 71, Mar. 2005, pp. 1291-1299. |
 "
"