Team:KULeuven/Model/MultiCell
From 2008.igem.org
(→Multi-cell Modeling) |
m |
||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
| + | {{:Team:KULeuven/Tools/Scripting}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| - | |||
| - | |||
== Multi-cell Modeling == | == Multi-cell Modeling == | ||
| - | The output of our | + | [[Image:Multicell icon.PNG|right|300px]] |
| + | |||
| + | The output of our InverTimer, the LuxI protein, causes an accumulating amount of HSL molecules, which are in fact signaling molecules. This means that they are used by cells to communicate with each other by diffusing in the environment. | ||
At this point we have only simulated 1 Dr. Coli @ work. It might be a good idea to take this model to a higher level and simulate the behaviour of a colony of cells. At a later stage we can add a diffusive environment (medium), in which the signaling molecules can travel from one cell to a neighboring cell, and see what happens... | At this point we have only simulated 1 Dr. Coli @ work. It might be a good idea to take this model to a higher level and simulate the behaviour of a colony of cells. At a later stage we can add a diffusive environment (medium), in which the signaling molecules can travel from one cell to a neighboring cell, and see what happens... | ||
| Line 24: | Line 26: | ||
Second, we also implemented some functions to be able to simulate the whole diagram with events and some other features which are present in the Simbiology Toolbox. At this point we are able to run simulations with a growing (Dr. Coli) cell population while they are susceptible to external signals (TetR-input). | Second, we also implemented some functions to be able to simulate the whole diagram with events and some other features which are present in the Simbiology Toolbox. At this point we are able to run simulations with a growing (Dr. Coli) cell population while they are susceptible to external signals (TetR-input). | ||
| - | + | <html> | |
| + | <div class="center"> | ||
| + | <div class="noborder" style="overflow: auto; width: 800px; height: 420px;"> | ||
| + | <div class="noborder" style="width: 1250px;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/9/92/Multicell_HSL1.png" style="float: left; width: 600px; height: 400px; margin: 0 5px;" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/0/0f/Multicell_ccdB1.png" style="float: left; width: 600px; height: 400px; margin: 0 5px;" /> | ||
| + | </div></div></div></html> | ||
Latest revision as of 23:26, 29 October 2008
Multi-cell Modeling
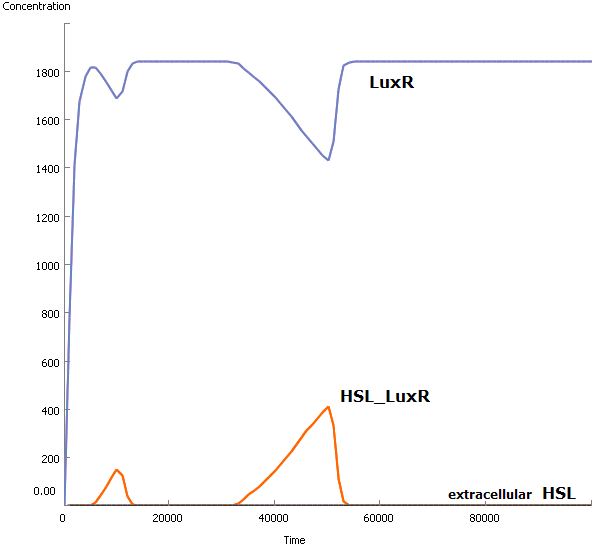
The output of our InverTimer, the LuxI protein, causes an accumulating amount of HSL molecules, which are in fact signaling molecules. This means that they are used by cells to communicate with each other by diffusing in the environment.
At this point we have only simulated 1 Dr. Coli @ work. It might be a good idea to take this model to a higher level and simulate the behaviour of a colony of cells. At a later stage we can add a diffusive environment (medium), in which the signaling molecules can travel from one cell to a neighboring cell, and see what happens...
Full Model
A small example leads to a simple conclusion: this analysis is unnecessary. Even when there is possible diffusion of HSL molecules to the environment, it won't happen because of the exces amount of LuxR molecules. LuxR will immediately bind the available HSL molecules (very high association rate) and no HSL will diffuse.
Full Model - Part 2
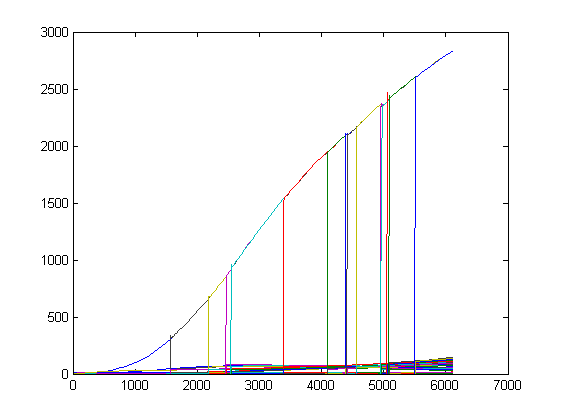
Because of the auto-inducible LuxR promotor there will certainly be diffusion into the medium (see Model - Part2): a deeper research needs to be done! Therefor we wrote a Matlab-script which is capable of interacting with the Simbiology Toolbox. This enables us to replicate cells in silico: we can simulate an increasing population of cells. A pretty visualisation is given below: each pop-up of a bar simulates the sudden presence of a new cell caused by the division of one of the available cells (starting population exists of 4 cells). This sudden pop-up effect is because we only simulate cell multiplication and not the growth of the new born cells: they are immediately at full size with about the same concentrations of the mother cell.
Second, we also implemented some functions to be able to simulate the whole diagram with events and some other features which are present in the Simbiology Toolbox. At this point we are able to run simulations with a growing (Dr. Coli) cell population while they are susceptible to external signals (TetR-input).


 "
"