Team:KULeuven/Model/Output
From 2008.igem.org
Contents |
Output
Position in the system
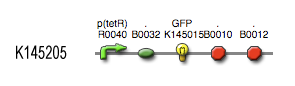
The output system is one of three modules directly linked to the output of the light sensor (black-box) the input for is OmpF. The output system is a simple gene regulation, of which transcription is activated by OmpF. The output signal is CFP (cyan fluorescent protein).
In the applied project of dealing with Crohn's disease this would be replaced by the drug necessary to cure the local inflammation. The drug concentration would be proportional to the amount of local inflammation sensed.
Describing the system
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dCFP (protein) | 1.05E-4 s-1 | [http://parts.mit.edu/igem07/index.php/ETHZ/Parameters link] | |
| dRNA_CFP (mRNA) | 0.0023 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| Transcription Rates | |||
| kCFP | OmpF dependent | ||
| Translation Rates | |||
| kCFP | 0.167 s-1 | RBS is B0032 (efficiency of 0.3) | |
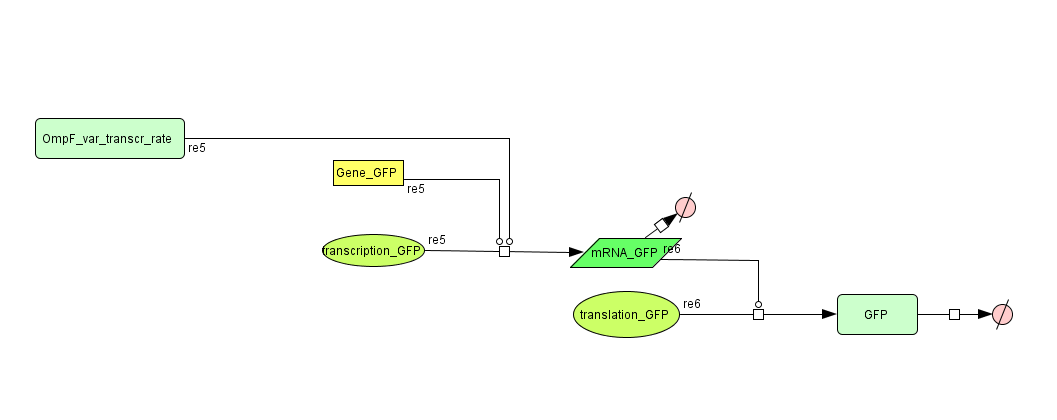
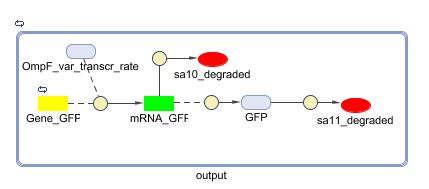
Models
CellDesigner (SBML file)
Matlab (SBML file)
Simulations
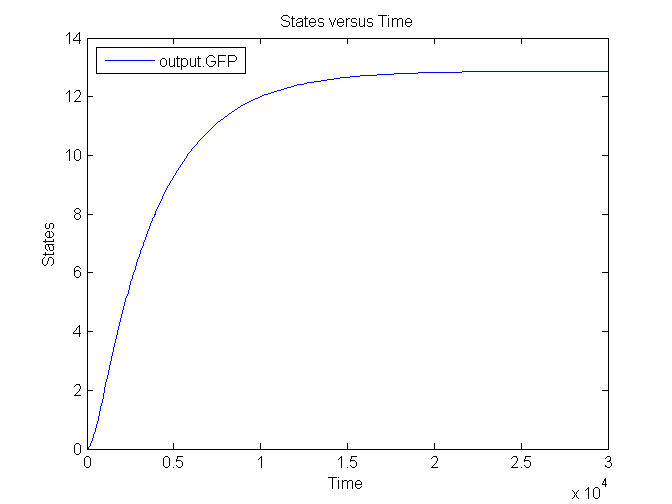
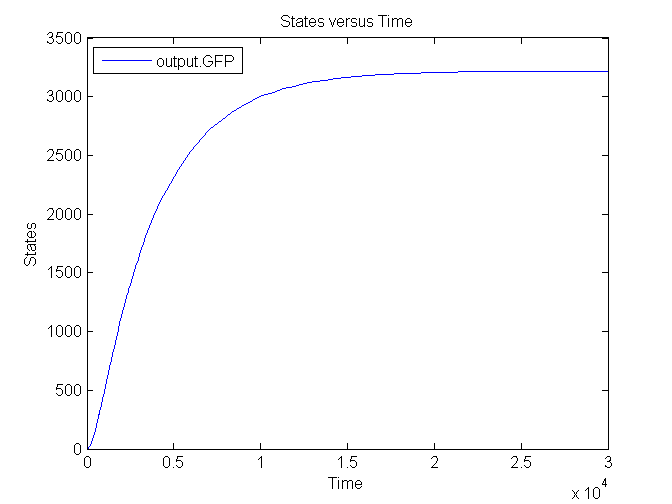
Several simulations were conducted, the following two show the transient respons and eventual saturation of the output system to a low and a high step signal as input, respectively. We see that there is a great distinction in the number of CFP molecules between the two input signals.
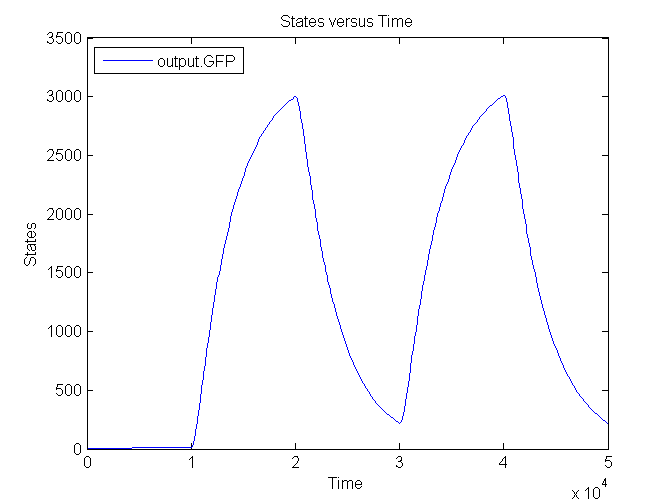
Next figure shows the switching behaviour of the output system to an light signal turning off and on:
 "
"