Team:KULeuven/Software/Simbiology2LaTeX
From 2008.igem.org
Contents |
Introduction
We've written a small toolbox to convert the information in the Matlab Simbiology models to LaTeX-code. This code can be easily converted to a pdf-file with various open source programs (e.g. [http://www.texniccenter.org/index.php?id=20 TeXnicCenter for Windows]). The idea for this toolbox is based on the plugin [http://systems-biology.org/cd/plugins.html Squeezer] for CellDesigner.
The Program
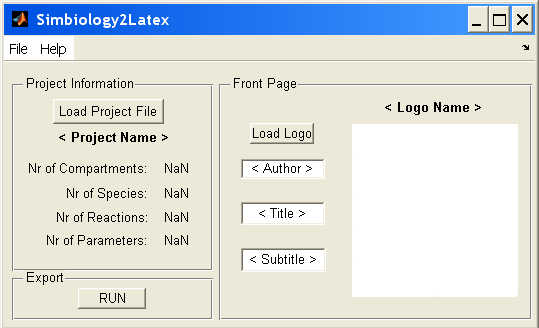
GUI
The program consists of GUI as interface between the user and the converting code. You can start the program by typing "Simbiology2Latex" in the Matlab console. The GUI looks like this:
Use
Step 1
The first thing you have to do is import the model. You can do this by clicking on the "Load Project File" button or by File > Load in the menu bar. After you imported the right model, you'll see some information about your project, like the project name, the number of compartments, reactions, ...
Step 2
The second step is optional and consists of different substeps. With the button "Load Logo" you can import a logo (e.g. the team logo) that will be on the front page of your pdf-file. You can also give the name of the author, title and subtitle of the project.
Step 3
Push "RUN" to generate your TeX-file.
Step 4
Open an Editor to convert the TeX-file into a pdf-file.
Problems
The formatted equations are sometimes too long. There can only be one model in the project file. The first model will always the chosen one.
 "
"