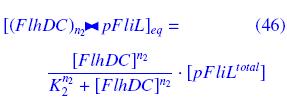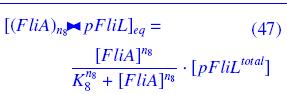Team:Paris/Modeling/FromMolReactToNLOde
From 2008.igem.org
(→Mathematical Interpretation of the Molecular Reactions) |
|||
| (31 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Paris/Header|From Molecular Reactions to Non-Linear ODEs}} | {{Paris/Header|From Molecular Reactions to Non-Linear ODEs}} | ||
| - | + | {{Paris/Section_contents_characterization}} | |
| - | + | ||
== Idea and Assumptions == | == Idea and Assumptions == | ||
| - | We will here propose a Mathematical Modeling of the | + | We will here propose a Mathematical Modeling of the [[Team:Paris/Modeling/Molecular_Reactions|elementary molecular reactions]]. The idea of the ''Characterization Approach'' is that this modelization must both accounts for [[Team:Paris/Modeling/Protocol_Of_Characterization|every small steps]] of the system and allow the [[Team:Paris/Modeling/Protocol_Of_Characterization|experimental characterizations]]. |
| - | |||
| - | |||
| - | |||
| - | Therefore, the following equations do not describe properly what ''really'' happens in the cells. For exemple, we know that the transcription factor FlhD-FlhC is actually an ''hexamere FlhD<sub>4</sub>C<sub>2</sub>''. But, as we will surely not get access to the ''dissociation constant'' of the ''hexamerisation'', we just treat it as an ''abstract'' inducer protein "''FlhDC''", with an order (''n'') in its ''complexation caracterization'' probably around 2*4 = 8 (but perhaps completly different ! ; the estimation of the error by the | + | <br> |
| + | |||
| + | Therefore, the following equations do not describe properly what ''really'' happens in the cells. For exemple, we know that the transcription factor FlhD-FlhC is actually an ''hexamere FlhD<sub>4</sub>C<sub>2</sub>''. But, as we will surely not get access to the ''dissociation constant'' of the ''hexamerisation'', we just treat it as an ''abstract'' inducer protein "''FlhDC''", with an order (''n'') in its ''complexation caracterization'' probably around 2*4 = 8 (but perhaps completly different ! ; the estimation of the error by the [[Team:Paris/Modeling/Implementation#Parameters_Finder_Programs|Parameters Finder Programs]] would tell us if it stays consistent). | ||
<div style="text-align: center"> | <div style="text-align: center"> | ||
| - | {{Paris/Toggle|all assumptions|Team:Paris/Modeling/More_From2Ode_Explanation}} | + | {{Paris/Toggle|all assumptions|Team:Paris/Modeling/More_From2Ode_Explanation|250px}} |
</div> | </div> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
== Mathematical Interpretation and Simulation of the Molecular Reactions == | == Mathematical Interpretation and Simulation of the Molecular Reactions == | ||
| - | First, we consider the '''complexation phenomenon'''. We show, under the | + | First, we consider the '''complexation phenomenon'''. We show, under the ''quasi steady-state hypothesis'', how it leads to ''Non-Linear interactions'' like "Hill functions". |
<div style="text-align: center"> | <div style="text-align: center"> | ||
| - | {{Paris/Toggle| | + | {{Paris/Toggle|developments|Team:Paris/Modeling/More_From2Ode_M|250px}} |
</div> | </div> | ||
| - | Then, we apply these results to the | + | <br> |
| + | |||
| + | Then, we apply these results to the ''Molecular Reactions'' of our system. That gives all the theoretical calculation of the complexations, and finally, with the ''previous assumptions'', we get the ''equations of the full system of ODEs''. | ||
{| border="1" width="100%" style="text-align: center" | {| border="1" width="100%" style="text-align: center" | ||
| Line 35: | Line 38: | ||
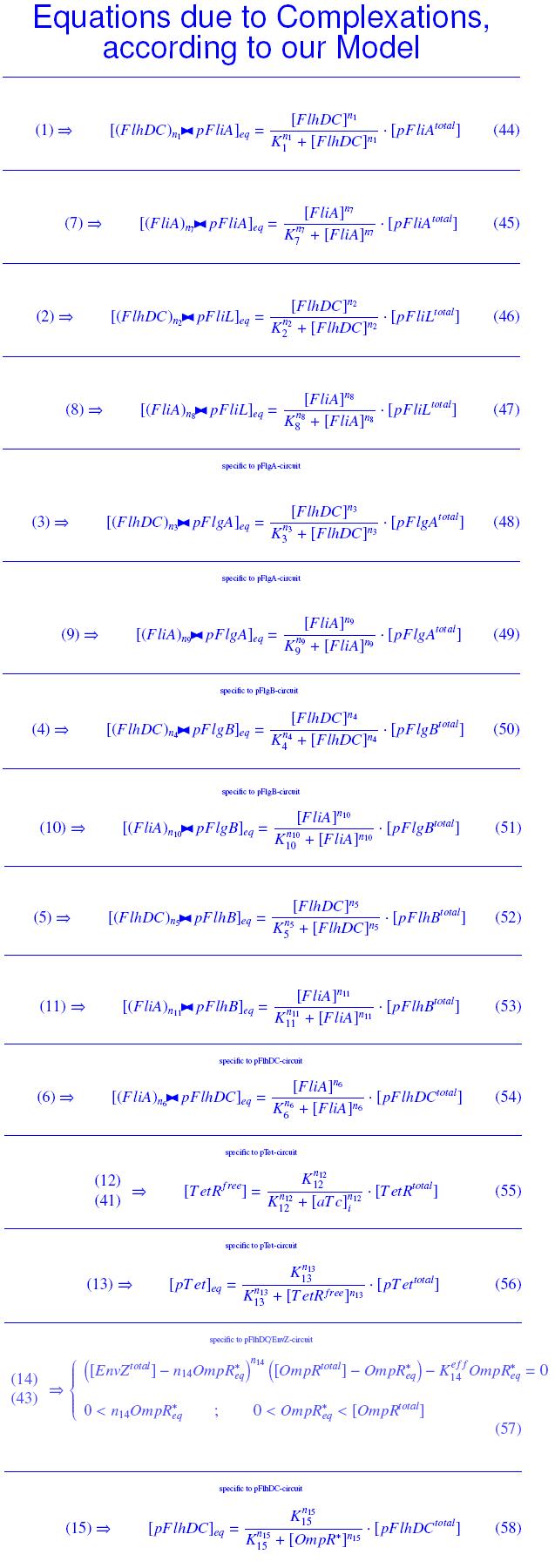
[[Image:compl.jpg|110px]] means ''n<sub>1</sub>'' molecules of ''FlhDC'' complexed with a ''pFliA'' promoter<br><br> | [[Image:compl.jpg|110px]] means ''n<sub>1</sub>'' molecules of ''FlhDC'' complexed with a ''pFliA'' promoter<br><br> | ||
| - | < | + | <div style="text-align: center"> |
| + | {{Paris/Toggle2|Complexation Steady-State|Team:Paris/Modeling/Compl_SS}} | ||
| + | </div> | ||
| width="25%" | '''On the Example :''' <br> | | width="25%" | '''On the Example :''' <br> | ||
<small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small><br> | <small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small><br> | ||
<br> | <br> | ||
| - | * ''FlhDC'' and ''FliA'' bind to ''pFliL'' : <br> | + | * ''FlhDC'' and ''FliA'' bind to ''pFliL''<br> |
| - | [[Image:DCiLcomplEq.jpg]] | + | following "Hill functions" : <br> |
| + | [[Image:DCiLcomplEq.jpg|270px]] | ||
| - | [[Image:AiLcomplEq.jpg]] | + | [[Image:AiLcomplEq.jpg|270px]] |
|- | |- | ||
| Line 50: | Line 56: | ||
[[Image:compl.jpg|110px]] means ''n<sub>1</sub>'' molecules of ''FlhDC'' complexed with a ''pFliA'' promoter<br><br> | [[Image:compl.jpg|110px]] means ''n<sub>1</sub>'' molecules of ''FlhDC'' complexed with a ''pFliA'' promoter<br><br> | ||
| - | < | + | <div style="text-align: center"> |
| + | {{Paris/Toggle2|ODEs of the Global System|Team:Paris/Modeling/ODEs}} | ||
| + | </div> | ||
| width="25%" | '''On the Example :''' <br> | | width="25%" | '''On the Example :''' <br> | ||
| - | <small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small | + | <small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small> |
<br> | <br> | ||
| - | * ''FlhDC><pFliL'' and ''FliA><pFliL'' causes "production" of ''FP1'', | + | * ''FlhDC><pFliL'' and ''FliA><pFliL'' causes "production" of ''FP1'', diluted along times : |
| - | + | <br> | |
| - | [[Image:bFP1dot.jpg]] | + | [[Image:bFP1dot.jpg|bFP1dot.jpg]] |
| - | + | the outlined parameters are necessary and enough to fully characterize ƒ5 | |
|} | |} | ||
| + | <div style="text-align: right">For complete PDF with all Reactions and Numbered References : [https://static.igem.org/mediawiki/2008/6/61/Equa_Core_System.pdf complete list of reactions and equations] </div> | ||
| + | |||
<br> | <br> | ||
| - | + | ||
| + | {{Paris/Navig|Team:Paris/Modeling/Workflow_Example}} | ||
Latest revision as of 03:39, 30 October 2008
|
From Molecular Reactions to Non-Linear ODEs
Other pages:
Idea and AssumptionsWe will here propose a Mathematical Modeling of the elementary molecular reactions. The idea of the Characterization Approach is that this modelization must both accounts for every small steps of the system and allow the experimental characterizations.
Therefore, the following equations do not describe properly what really happens in the cells. For exemple, we know that the transcription factor FlhD-FlhC is actually an hexamere FlhD4C2. But, as we will surely not get access to the dissociation constant of the hexamerisation, we just treat it as an abstract inducer protein "FlhDC", with an order (n) in its complexation caracterization probably around 2*4 = 8 (but perhaps completly different ! ; the estimation of the error by the Parameters Finder Programs would tell us if it stays consistent). ↓ all assumptions ↑
Mathematical Interpretation and Simulation of the Molecular ReactionsFirst, we consider the complexation phenomenon. We show, under the quasi steady-state hypothesis, how it leads to Non-Linear interactions like "Hill functions". ↓ developments ↑
Then, we apply these results to the Molecular Reactions of our system. That gives all the theoretical calculation of the complexations, and finally, with the previous assumptions, we get the equations of the full system of ODEs. For complete PDF with all Reactions and Numbered References : complete list of reactions and equations
|
 "
"