Team:UC Berkeley/Assembly
From 2008.igem.org
ChrisBrown (Talk | contribs) |
ChrisBrown (Talk | contribs) |
||
| Line 8: | Line 8: | ||
=='''Assembly in Cell Lysate'''== | =='''Assembly in Cell Lysate'''== | ||
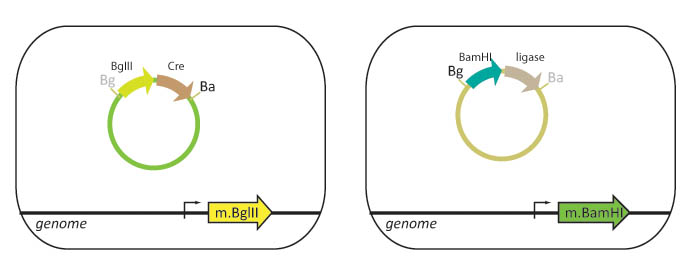
| - | Our lab currently | + | Our lab currently uses cells that are engineered to methylate either BamHI or BglII restriction sites. Part A is transformed into a "lefty" cell that is methylated on BglII restriction sites while Part B is transformed into a "righty" cell that is methylated on BamHI restriction sites. |
We propose to integrate the BamHI and BglII genes into the ''E. coli'' genome. In this scheme, lefty cells that are methylated on BglII cut sites, will stably express ligase and BglII. Righty cells that are methylated on BamHI cut sites, will be engineered to stably express XhoI and BamHI. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The methylation also protects the cellular DNA from being cut when these genes are expressed. | We propose to integrate the BamHI and BglII genes into the ''E. coli'' genome. In this scheme, lefty cells that are methylated on BglII cut sites, will stably express ligase and BglII. Righty cells that are methylated on BamHI cut sites, will be engineered to stably express XhoI and BamHI. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The methylation also protects the cellular DNA from being cut when these genes are expressed. | ||
| - | + | We propose to eliminate the need for mini-prep by using our lysis device [[https://2008.igem.org/Team:UC_Berkeley/LysisDevice]] and the BamHI/BglII/XhoI/ligase cells to lyse the cells and release the restriction enzymes and ligase into the lysate. The lysate mixture is incubated to allow time for assembly (digestion and ligation). The lysate is then used to transform cells and cells are plated on the appropriate antibiotic. | |
| - | + | ||
| - | + | ||
[[Image:cdb6.jpg]] | [[Image:cdb6.jpg]] | ||
| - | + | ==='''Testing and Experimentation'''=== | |
| - | + | A. To test the viablity of the plasmid released by the lysis device - | |
| - | We propose to engineer cells that are | + | Cells containing basic part plasmid DNA were lysed with the lysis device. The lysate was used to transform another batch of cells. This experiment was produced many colonies. |
| + | |||
| + | [[Image:lysed_cells.jpg]] | ||
| + | |||
| + | B. To test the viability of enzymes in the lysate - | ||
| + | |||
| + | Lefty and righty cells were combined in a single eppendorf tube and lysed with our lysis device. Commercial restriction enzymes and ligase were added to the lysate along with plasmid DNA. The mixture was incubated. The lysate was used to transform competant cells. The comp cells were plated on the appropriate antibiotic, but failed to produce colonies. | ||
| + | |||
| + | The experiment was repeated with lysed cells that were centrifuged and re-suspended in Buffer NEB2. This experiment produced the colonies with the correct composite part when plated on the appropriate antibiotic. | ||
| + | |||
| + | C. Testing digestion in lysate | ||
| + | |||
| + | Lefty and righty cells containing plasmid DNA were lysed with our lysis device. Lysed cells were centrifuged and the supernatant was discarded. Cells were re-suspended in NEB2 Buffer. Commercial restriction enzymes and ligase were added to the lysate and the lysate was incubated. The lysate was used then used to transform competant cells. | ||
| + | |||
| + | The exact conditions required to make this experiment successful is difficult to determine. Since the lysis device results in successful release of plasmid DNA and assembly works in NEB2 buffered lysate, digestion of plasmid DNA in the lysate should work under the appropriate conditions. However, at the present, this experiment does not produce the correct colonies. | ||
| + | |||
| + | D. Testing the viability of enzymes produced by the cells | ||
| + | |||
| + | 1) Ligase strain – The gene for ligase has been cloned into the genome of lefty and righty cells. These strains have successfully been used to optimize the effectiveness of the ligation reaction. | ||
| + | |||
| + | 2) Bam strain - The gene for the BamHI restriction enzyme has been cloned into cells [[insert data here]] | ||
| + | |||
| + | =='''Assembly in-Vivo using Phagemid'''== | ||
| + | |||
| + | We propose to engineer assembler cells that are express BamHI methylase and BglII methylase and stably express BamHI, BglII, Cre and ligase. The assembler cell contains all of the genes needed for the lytic cycle of the phage. The three genes necessary to induce the lytic cycle of the phage are initially repressed. | ||
| + | |||
| + | Bacteriophages with phagemids containing a basic part flanked by BglII and BamHI restriction sites and two antibiotic resistance genes separated by a XhoI restriction site are created. To methylate phagemid DNA, lefty and righty cells will be infected with these phages. | ||
| + | |||
| + | Assembler cells will be infected with both lefty and righty phagemids. The cells produce the restriction enzymes and ligase necessary to complete an in-vivo assembly. The lytic cycle of the phage is induced by removing the repressor on the lytic genes. The cells are lysed and phagemids are released into solution. The lysate is used to infect new cells and screened by plating on the appropriate antibiotic. | ||
| + | |||
| + | [[Image:Phagemid_assembly]] | ||
Revision as of 06:44, 29 October 2008
Our goal is to simplify two antibiotic assembly eliminating the replace mini-prep steps and making the protocol reagent-free. This will be accomplished by using our lysis device to lyse cells and extract DNA and engineering cells to produce their own restriction enzymes and ligase.
For a general overview of two antibiotic assembly, click here [[1]]
Assembly in Cell Lysate
Our lab currently uses cells that are engineered to methylate either BamHI or BglII restriction sites. Part A is transformed into a "lefty" cell that is methylated on BglII restriction sites while Part B is transformed into a "righty" cell that is methylated on BamHI restriction sites.
We propose to integrate the BamHI and BglII genes into the E. coli genome. In this scheme, lefty cells that are methylated on BglII cut sites, will stably express ligase and BglII. Righty cells that are methylated on BamHI cut sites, will be engineered to stably express XhoI and BamHI. Since restriction enzymes will not cut methylated DNA, the BglII restriction site in the lefty cell and the BamHI restriction site in the righty cell are blocked from digestion. The methylation also protects the cellular DNA from being cut when these genes are expressed.
We propose to eliminate the need for mini-prep by using our lysis device [[2]] and the BamHI/BglII/XhoI/ligase cells to lyse the cells and release the restriction enzymes and ligase into the lysate. The lysate mixture is incubated to allow time for assembly (digestion and ligation). The lysate is then used to transform cells and cells are plated on the appropriate antibiotic.
Testing and Experimentation
A. To test the viablity of the plasmid released by the lysis device -
Cells containing basic part plasmid DNA were lysed with the lysis device. The lysate was used to transform another batch of cells. This experiment was produced many colonies.
B. To test the viability of enzymes in the lysate -
Lefty and righty cells were combined in a single eppendorf tube and lysed with our lysis device. Commercial restriction enzymes and ligase were added to the lysate along with plasmid DNA. The mixture was incubated. The lysate was used to transform competant cells. The comp cells were plated on the appropriate antibiotic, but failed to produce colonies.
The experiment was repeated with lysed cells that were centrifuged and re-suspended in Buffer NEB2. This experiment produced the colonies with the correct composite part when plated on the appropriate antibiotic.
C. Testing digestion in lysate
Lefty and righty cells containing plasmid DNA were lysed with our lysis device. Lysed cells were centrifuged and the supernatant was discarded. Cells were re-suspended in NEB2 Buffer. Commercial restriction enzymes and ligase were added to the lysate and the lysate was incubated. The lysate was used then used to transform competant cells.
The exact conditions required to make this experiment successful is difficult to determine. Since the lysis device results in successful release of plasmid DNA and assembly works in NEB2 buffered lysate, digestion of plasmid DNA in the lysate should work under the appropriate conditions. However, at the present, this experiment does not produce the correct colonies.
D. Testing the viability of enzymes produced by the cells
1) Ligase strain – The gene for ligase has been cloned into the genome of lefty and righty cells. These strains have successfully been used to optimize the effectiveness of the ligation reaction.
2) Bam strain - The gene for the BamHI restriction enzyme has been cloned into cells insert data here
Assembly in-Vivo using Phagemid
We propose to engineer assembler cells that are express BamHI methylase and BglII methylase and stably express BamHI, BglII, Cre and ligase. The assembler cell contains all of the genes needed for the lytic cycle of the phage. The three genes necessary to induce the lytic cycle of the phage are initially repressed.
Bacteriophages with phagemids containing a basic part flanked by BglII and BamHI restriction sites and two antibiotic resistance genes separated by a XhoI restriction site are created. To methylate phagemid DNA, lefty and righty cells will be infected with these phages.
Assembler cells will be infected with both lefty and righty phagemids. The cells produce the restriction enzymes and ligase necessary to complete an in-vivo assembly. The lytic cycle of the phage is induced by removing the repressor on the lytic genes. The cells are lysed and phagemids are released into solution. The lysate is used to infect new cells and screened by plating on the appropriate antibiotic.
 "
"