Team:UC Berkeley/LayeredAssembly
From 2008.igem.org
ChrisBrown (Talk | contribs) |
|||
| Line 2: | Line 2: | ||
==Project Motivation== | ==Project Motivation== | ||
| - | + | ====Project Overview==== | |
| - | Our efforts to optimize synthetic biology protocols centered around the | + | Imagine a world with no mini-preps. A world where DNA extraction and purification could be accomplished in a single eppendorf tube. We did, and so we created CloneBots, an automated approach to synthetic biology. |
| + | |||
| + | CloneBots seeks to simplify the creation of composite parts to entail only liquid handling steps that could more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the ''E. coli'' genome, thereby allowing them to perform the required protocols in-vivo. | ||
| + | |||
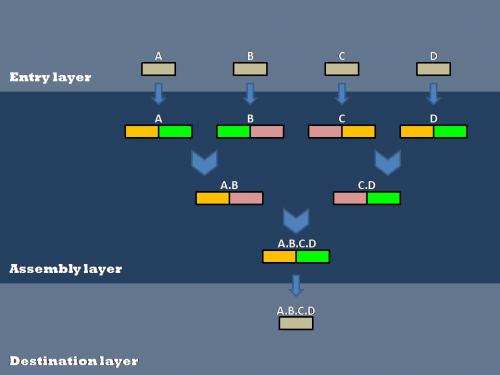
| + | Our efforts to optimize synthetic biology protocols centered around the layered assembly scheme. This involves the creation of biobrick parts in an entry vector, their transfer into double antibiotic assembly vectors(antibiotic resistances shown in orange, green, and pink), and subsequent assembly steps to connect various parts(A, B, C, and D). After all of the assembly steps are complete, the desired composite parts can then be transferred into the destination vector of choice. | ||
[[Image:layered Assembly.jpg|center|frame|350px|A schematic representation of Layered Assembly]] | [[Image:layered Assembly.jpg|center|frame|350px|A schematic representation of Layered Assembly]] | ||
| + | |||
| + | Layered assembly is a straight-forward and robust method for combining two or more basic parts into a destination vector. There are three layers (Entry, Assembly and Destination). Both Entry and Assembly layers are standardized for ease of use, while the Destination plasmid can be tailored to a specific experiment. | ||
| + | |||
| + | ====Entry Layer==== | ||
| + | |||
| + | The Entry plasmid contains a Spectinomycin antibiotic resistance marker and EcoRI, BglII and BamHI restriction sites. All basic parts are cloned into the entry vector. The basic part is flanked by attR1 and attr2 recombination sites. | ||
| + | |||
| + | Since the entry plasmid attR1 and attR2 recombination sites, the basic part can easily be transferred into one of six different assembly vectors using the Gateway cloning scheme described here [[https://2008.igem.org/Team:UC_Berkeley/GatewayOverview]]. | ||
| + | |||
| + | ====Assembly Layer==== | ||
| + | |||
| + | The assembly plasmid contains two antibiotic resistance genes, carefully chosen to simplify screening for the correct composite part. For details on the 2 antibiotic assembly scheme, click here [[https://2008.igem.org/Team:UC_Berkeley/Assembly]]. | ||
| + | |||
| + | ====Destination Layer==== | ||
| + | |||
| + | Completed composite parts are transferred from the assembly layer to the destination layer using the Gateway cloning scheme [[https://2008.igem.org/Team:UC_Berkeley/GatewayOverview]]. The Destination plasmid is tailored to a specific experiment or assay. This allows a single composite part to be transfered to several different Destination plasmids for further experimentation or characterization. | ||
Revision as of 04:19, 28 October 2008
Contents |
Project Motivation
Project Overview
Imagine a world with no mini-preps. A world where DNA extraction and purification could be accomplished in a single eppendorf tube. We did, and so we created CloneBots, an automated approach to synthetic biology.
CloneBots seeks to simplify the creation of composite parts to entail only liquid handling steps that could more readily be performed by robots. We have accomplished this by genetically encoding many of the required steps into the E. coli genome, thereby allowing them to perform the required protocols in-vivo.
Our efforts to optimize synthetic biology protocols centered around the layered assembly scheme. This involves the creation of biobrick parts in an entry vector, their transfer into double antibiotic assembly vectors(antibiotic resistances shown in orange, green, and pink), and subsequent assembly steps to connect various parts(A, B, C, and D). After all of the assembly steps are complete, the desired composite parts can then be transferred into the destination vector of choice.
Layered assembly is a straight-forward and robust method for combining two or more basic parts into a destination vector. There are three layers (Entry, Assembly and Destination). Both Entry and Assembly layers are standardized for ease of use, while the Destination plasmid can be tailored to a specific experiment.
Entry Layer
The Entry plasmid contains a Spectinomycin antibiotic resistance marker and EcoRI, BglII and BamHI restriction sites. All basic parts are cloned into the entry vector. The basic part is flanked by attR1 and attr2 recombination sites.
Since the entry plasmid attR1 and attR2 recombination sites, the basic part can easily be transferred into one of six different assembly vectors using the Gateway cloning scheme described here [[1]].
Assembly Layer
The assembly plasmid contains two antibiotic resistance genes, carefully chosen to simplify screening for the correct composite part. For details on the 2 antibiotic assembly scheme, click here [[2]].
Destination Layer
Completed composite parts are transferred from the assembly layer to the destination layer using the Gateway cloning scheme [[3]]. The Destination plasmid is tailored to a specific experiment or assay. This allows a single composite part to be transfered to several different Destination plasmids for further experimentation or characterization.
 "
"