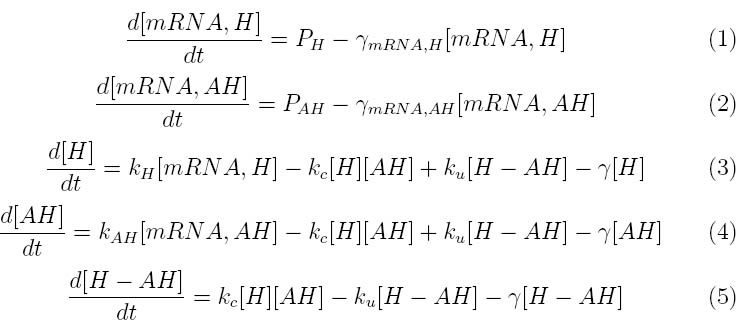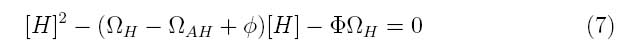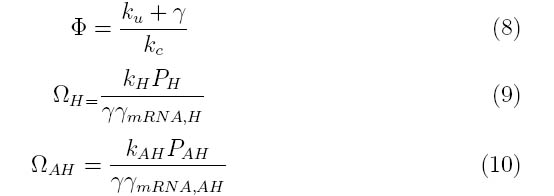Team:UC Berkeley/Modeling
From 2008.igem.org
ChrisBrown (Talk | contribs) |
ChrisBrown (Talk | contribs) |
||
| Line 19: | Line 19: | ||
[[Image:cdb3.jpg]] | [[Image:cdb3.jpg]] | ||
| - | Where | + | Where the system can be divided into three dimensionless parameters which describe the behavior of holin, antiholin and the holin-antiholin dimer |
[[Image:cdb4.jpg]] | [[Image:cdb4.jpg]] | ||
| - | Using MatLab, | + | Using MatLab, the following graph of the system was produced. |
[[Image:image1.png]] | [[Image:image1.png]] | ||
| + | |||
| + | The horizontal line at y=1 represents the critical holin concentration needed induce lysis. | ||
| + | |||
[[Image:image2.png]] | [[Image:image2.png]] | ||
[[Image:image3.png]] | [[Image:image3.png]] | ||
Revision as of 05:41, 25 October 2008
The kinetics of the Lambda phage lysis device was modeled to help our team gain an insight into the behavior of our system. The below equations describe our system
Where P1 and P2 approximate the mRNA promoter strength and /omega_mH and mH are the degradation rates for holin and antiholin respectively. kH , kAH and kc represent the rate constants for holin and antiholin formation and the coupling rate for the holin-antiholin dimer.
The literature indicates that at the time of lysis, cells infected with lambda phage have approximately 1000 holin proteins. Therefore, the critical concentration of holin (Hc) was set at 1000 holin proteins per cell.
At steady state,
The system can be simplified into the following transfer function
Where the system can be divided into three dimensionless parameters which describe the behavior of holin, antiholin and the holin-antiholin dimer
Using MatLab, the following graph of the system was produced.
The horizontal line at y=1 represents the critical holin concentration needed induce lysis.
 "
"