Team:UNIPV-Pavia/Notebook/Week12
From 2008.igem.org
(Difference between revisions)
| (15 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
!|[[Team:UNIPV-Pavia/Notebook/Week13|Week 13]] | !|[[Team:UNIPV-Pavia/Notebook/Week13|Week 13]] | ||
!|[[Team:UNIPV-Pavia/Notebook/Week14|Week 14]] | !|[[Team:UNIPV-Pavia/Notebook/Week14|Week 14]] | ||
| + | |- | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week15|Week 15]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week16|Week 16]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week17|Week 17]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week18|Week 18]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week19|Week 19]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week20|Week 20]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week21|Week 21]] | ||
| + | |- | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week22|Week 22]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week23|Week 23]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week24|Week 24]] | ||
|} | |} | ||
<br><br> | <br><br> | ||
| - | ==Week 11: 08/4/08 - 08/ | + | ==Week 11: 08/4/08 - 08/7/08== |
'''08/4/08''' | '''08/4/08''' | ||
| Line 48: | Line 60: | ||
*Plasmid digestion for: | *Plasmid digestion for: | ||
{|cellpadding="20px" | {|cellpadding="20px" | ||
| - | | | + | |J23100-B0030-C0040-'''B1006''' (E-S) |
| - | | | + | |R0040 (E-X) |
|} | |} | ||
*Gel run/cut/gel extraction. | *Gel run/cut/gel extraction. | ||
| - | *Ligation: | + | *Ligation: J23100-B0030-C0040-B1006-'''R0040'''. We incubated ligation at 16°C overnight. |
*We had 5 plates to screen with colony PCR: | *We had 5 plates to screen with colony PCR: | ||
| - | ** | + | **B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006 (that we call "a") |
| - | ** | + | **B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 (that we call "b") |
| - | ** | + | **B0030-C0078-B1006-'''R0079'''-B0030-E0040-B1006 (that we call "c") |
| - | ** | + | **B0030-C0061-B0030-C0079-B1006-'''R0079'''-B0030-E0040-B1006 (that we call "d") |
| - | ** | + | **J23100-B0030-C0012-B1006-'''R0010''' |
| - | *Last week | + | *Last week J23100-B0030-C0012-B1006-'''R0010''' colony PCR gave a bad result. For this reason, we decided to perform colony PCR only for: |
| - | ** | + | **B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 (7 colonies) |
| - | ** | + | **J23100-B0030-C0012-B1006-'''R0010''' (6 colonies) |
{| | {| | ||
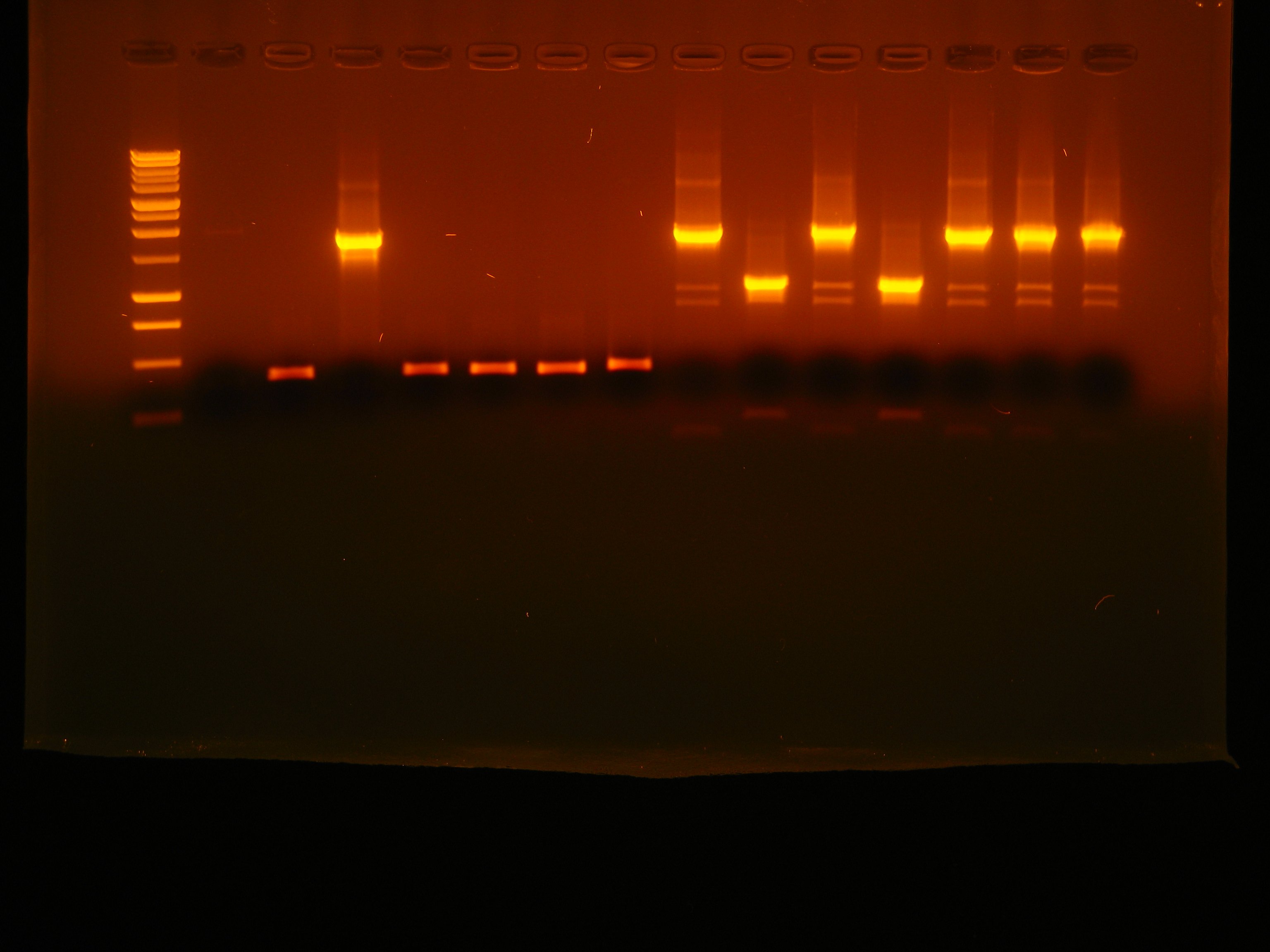
| - | |[[Image:pv_colonypcr_27_b.jpg|thumb| | + | |[[Image:pv_colonypcr_27_b.jpg|thumb|370px|left|Marker 1Kb, blank, J23100-B0030-C0012-B1006-'''R0010''' (6 colonies), B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 (7 colonies)]] |
|} | |} | ||
*Gel result: | *Gel result: | ||
| - | ** | + | **B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 (1st colony, but it was not pure. We decided to prepare single colonies plate for it) |
| - | ** | + | **J23100-B0030-C0012-B1006-'''R0010''' (2nd colony) |
'''08/4/08''' | '''08/4/08''' | ||
<br> | <br> | ||
| - | * | + | *We transformed J23100-B0030-C0040-B1006-'''R0040''' overnight ligation. We plated transformed bacteria and incubated plate at 37°C overnight. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | * | + | *We infected 9 ml of LB + Amp with 30 µl of C0062, Lig.12, Lig.22, Lig.30, Lig.27(2nd col), B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 (1st col) (see "Parts" section for our nomenclature). |
| - | * | + | *Single colonies plates for: |
| + | **B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 ("Lig.b") | ||
| + | **B0030-C0078-B1006-'''R0079'''-B0030-E0040-B1006 ("Lig.c") | ||
| + | **B0030-C0061-B0030-C0079-B1006-'''R0079'''-B0030-E0040-B1006 ("Lig.d") | ||
| - | + | '''08/5/08''' | |
| - | + | ||
| - | '''08/ | + | |
<br> | <br> | ||
| - | *Glycerol stocks/miniprep for | + | *Glycerol stocks/miniprep for C0062, Lig.12, Lig.22, Lig.30, Lig.27(2nd col), B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006(1). |
| - | *We sent | + | *We sent C0062, Lig.12, Lig.22 and Lig.30 purified plasmids to Primm for sequencing: all these parts contain BBa_C0062. |
| - | *We transformed/plated | + | *We transformed/plated J23100-B0030-C0040-B1006-'''R0040''' overnight ligation. |
| - | *Colony PCR for a(7 colonies), b(single colonies)(6 colonies), c(single colonies)(6 colonies), d(single colonies)(6 colonies). | + | *Colony PCR for a (7 colonies), Lig.b(single colonies)(6 colonies), Lig.c(single colonies)(6 colonies), Lig.d(single colonies)(6 colonies). |
| + | |||
| + | {| | ||
| + | |[[Image:pv_colonypcr_abcd.jpg|thumb|370px|left|Marker 1Kb, blank, Lig.a (7 colonies), Lig.b (6 colonies), Marker 1Kb, Lig.c (6 colonies), Lig.d (6 colonies)]] | ||
| + | |} | ||
*Gel results were not so clear: the length of some fragments was not expected and there were some contaminants. Maybe those parts were too long for our PCR reaction. We decided to grow 9 ml cultures for some of those colonies, to extract plasmids, to cut them and to check their length in a new run. We chose: | *Gel results were not so clear: the length of some fragments was not expected and there were some contaminants. Maybe those parts were too long for our PCR reaction. We decided to grow 9 ml cultures for some of those colonies, to extract plasmids, to cut them and to check their length in a new run. We chose: | ||
| - | ** | + | **B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006 (1, 4, 6, 7) |
| - | ** | + | **B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006 (no colony was chosen: we already had them and this run didn't show any 100% pure colony) |
| - | ** | + | **B0030-C0078-B1006-'''R0079'''-B0030-E0040-B1006 (5) |
| - | ** | + | **B0030-C0061-B0030-C0079-B1006-'''R0079'''-B0030-E0040-B1006 (2) |
| - | '''08/ | + | '''08/6/08''' |
<br> | <br> | ||
| - | *Single colonies plate for | + | *Single colonies plate for J23100-B0030-C0040-B1006-'''R0040''', because where were too many bacteria on its plate. |
*Glycerol stocks/miniprep for: | *Glycerol stocks/miniprep for: | ||
| - | ** | + | **B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(1) |
| - | ** | + | **B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(4) |
| - | ** | + | **B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(6) |
| - | ** | + | **B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(7) |
| - | ** | + | **B0030-C0078-B1006-'''R0079'''-B0030-E0040-B1006(5) |
| - | ** | + | **B0030-C0061-B0030-C0079-B1006-'''R0079'''-B0030-E0040-B1006(2) |
| - | *We cut these 6 plasmids E-P (5 µl) | + | *We cut these 6 plasmids and Lig.b in (E-P) (5 µl of DNA in a final reaction volume of 20 µl). |
| + | |||
| + | *Run for digested plasmids. | ||
| + | |||
| + | {| | ||
| + | |[[Image:pv_colonypcr_aaaabcd.jpg|thumb|370px|left|Marker 1Kb, Lig.a (1), Lig.a (4), Lig.a (6), Lig.a (7), Lig.b (1), Lig.c (5), Lig.d (2)]] | ||
| + | |} | ||
| + | |||
| + | *Gel results: | ||
| + | **(Lig.a) B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(1) OK | ||
| + | **(Lig.a) B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(4) False positive | ||
| + | **(Lig.a) B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(6) OK | ||
| + | **(Lig.a) B0030-C0051-B0030-C0079-'''B1006'''-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(7) False positive | ||
| + | **(Lig.b) B0030-C0061-B1006-'''R0062'''-B0030-E0040-B1006(1) OK | ||
| + | **(Lig.c) B0030-C0078-B1006-'''R0079'''-B0030-E0040-B1006(5) OK | ||
| + | **(Lig.d) B0030-C0061-B0030-C0079-B1006-'''R0079'''-B0030-E0040-B1006(2) OK | ||
| + | |||
| + | *We 9 ml of LB + Amp with 30 ul of Lig.b(1) and Lig.c(5) to perform tests. | ||
| + | |||
| + | '''08/7/08''' | ||
| + | <br> | ||
| + | *Qualitative fluorescence tests for Lig.b and Lig.c. Results are shown in The Project section (Experiments). | ||
| + | |||
| + | *Colony PCR for J23100-B0030-C0040-B1006-'''R0040''' single colonies plate (screening on 6 colonies). | ||
| + | |||
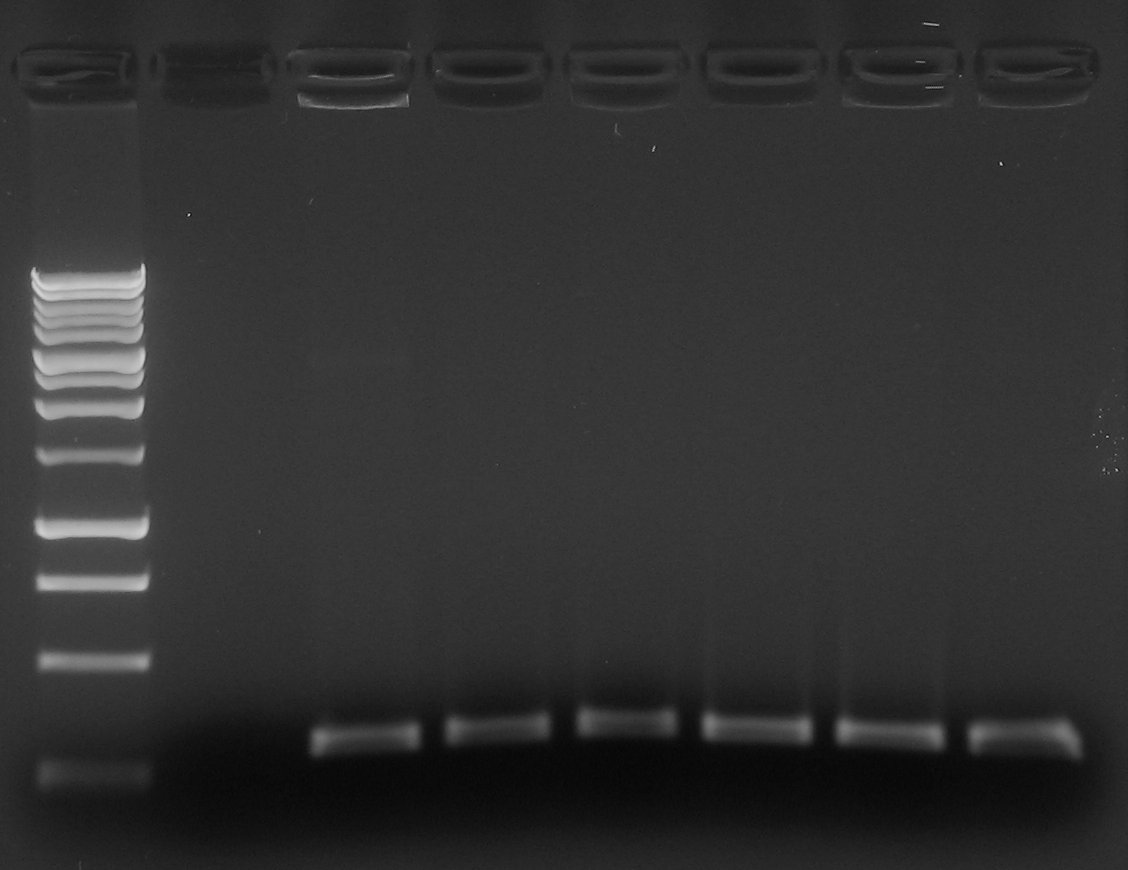
| + | *Gel results: all screened colonies were negative... | ||
| + | {| | ||
| + | |[[Image:pv_colonypcr_28_allnegatives.jpg|thumb|300px|left|Marker 1Kb, blank, J23100-B0030-C0040-B1006-'''R0040''' (6 colonies)]] | ||
| + | |} | ||
Latest revision as of 21:27, 26 October 2008
Notebook
| Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 |
|---|---|---|---|---|---|---|
| Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 14 |
| Week 15 | Week 16 | Week 17 | Week 18 | Week 19 | Week 20 | Week 21 |
| Week 22 | Week 23 | Week 24 |
Week 11: 08/4/08 - 08/7/08
08/4/08
- Plasmid digestion for:
| J23100-B0030-C0040-B1006 (E-S) | R0040 (E-X) |
- Gel run/cut/gel extraction.
- Ligation: J23100-B0030-C0040-B1006-R0040. We incubated ligation at 16°C overnight.
- We had 5 plates to screen with colony PCR:
- B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006 (that we call "a")
- B0030-C0061-B1006-R0062-B0030-E0040-B1006 (that we call "b")
- B0030-C0078-B1006-R0079-B0030-E0040-B1006 (that we call "c")
- B0030-C0061-B0030-C0079-B1006-R0079-B0030-E0040-B1006 (that we call "d")
- J23100-B0030-C0012-B1006-R0010
- Last week J23100-B0030-C0012-B1006-R0010 colony PCR gave a bad result. For this reason, we decided to perform colony PCR only for:
- B0030-C0061-B1006-R0062-B0030-E0040-B1006 (7 colonies)
- J23100-B0030-C0012-B1006-R0010 (6 colonies)
- Gel result:
- B0030-C0061-B1006-R0062-B0030-E0040-B1006 (1st colony, but it was not pure. We decided to prepare single colonies plate for it)
- J23100-B0030-C0012-B1006-R0010 (2nd colony)
08/4/08
- We transformed J23100-B0030-C0040-B1006-R0040 overnight ligation. We plated transformed bacteria and incubated plate at 37°C overnight.
- We infected 9 ml of LB + Amp with 30 µl of C0062, Lig.12, Lig.22, Lig.30, Lig.27(2nd col), B0030-C0061-B1006-R0062-B0030-E0040-B1006 (1st col) (see "Parts" section for our nomenclature).
- Single colonies plates for:
- B0030-C0061-B1006-R0062-B0030-E0040-B1006 ("Lig.b")
- B0030-C0078-B1006-R0079-B0030-E0040-B1006 ("Lig.c")
- B0030-C0061-B0030-C0079-B1006-R0079-B0030-E0040-B1006 ("Lig.d")
08/5/08
- Glycerol stocks/miniprep for C0062, Lig.12, Lig.22, Lig.30, Lig.27(2nd col), B0030-C0061-B1006-R0062-B0030-E0040-B1006(1).
- We sent C0062, Lig.12, Lig.22 and Lig.30 purified plasmids to Primm for sequencing: all these parts contain BBa_C0062.
- We transformed/plated J23100-B0030-C0040-B1006-R0040 overnight ligation.
- Colony PCR for a (7 colonies), Lig.b(single colonies)(6 colonies), Lig.c(single colonies)(6 colonies), Lig.d(single colonies)(6 colonies).
- Gel results were not so clear: the length of some fragments was not expected and there were some contaminants. Maybe those parts were too long for our PCR reaction. We decided to grow 9 ml cultures for some of those colonies, to extract plasmids, to cut them and to check their length in a new run. We chose:
- B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006 (1, 4, 6, 7)
- B0030-C0061-B1006-R0062-B0030-E0040-B1006 (no colony was chosen: we already had them and this run didn't show any 100% pure colony)
- B0030-C0078-B1006-R0079-B0030-E0040-B1006 (5)
- B0030-C0061-B0030-C0079-B1006-R0079-B0030-E0040-B1006 (2)
08/6/08
- Single colonies plate for J23100-B0030-C0040-B1006-R0040, because where were too many bacteria on its plate.
- Glycerol stocks/miniprep for:
- B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(1)
- B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(4)
- B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(6)
- B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(7)
- B0030-C0078-B1006-R0079-B0030-E0040-B1006(5)
- B0030-C0061-B0030-C0079-B1006-R0079-B0030-E0040-B1006(2)
- We cut these 6 plasmids and Lig.b in (E-P) (5 µl of DNA in a final reaction volume of 20 µl).
- Run for digested plasmids.
- Gel results:
- (Lig.a) B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(1) OK
- (Lig.a) B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(4) False positive
- (Lig.a) B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(6) OK
- (Lig.a) B0030-C0051-B0030-C0079-B1006-R0051-B0030-C0062-B1006-R0062-B0030-E1010-B1006(7) False positive
- (Lig.b) B0030-C0061-B1006-R0062-B0030-E0040-B1006(1) OK
- (Lig.c) B0030-C0078-B1006-R0079-B0030-E0040-B1006(5) OK
- (Lig.d) B0030-C0061-B0030-C0079-B1006-R0079-B0030-E0040-B1006(2) OK
- We 9 ml of LB + Amp with 30 ul of Lig.b(1) and Lig.c(5) to perform tests.
08/7/08
- Qualitative fluorescence tests for Lig.b and Lig.c. Results are shown in The Project section (Experiments).
- Colony PCR for J23100-B0030-C0040-B1006-R0040 single colonies plate (screening on 6 colonies).
- Gel results: all screened colonies were negative...
 "
"