Team:UNIPV-Pavia/Notebook/Week8
From 2008.igem.org
(Difference between revisions)
| Line 50: | Line 50: | ||
**BBa_J23100-'''BBa_B0030'''-BBa_I15010 (for the second time, we hoped to find true positive colonies) | **BBa_J23100-'''BBa_B0030'''-BBa_I15010 (for the second time, we hoped to find true positive colonies) | ||
**BBa_B0030-BBa_E0040-'''BBa_B1006''' | **BBa_B0030-BBa_E0040-'''BBa_B1006''' | ||
| - | **BBa_B0030-BBa_C0051-''' | + | **BBa_B0030-BBa_C0051-'''BBa_B0030''' |
**BBa_B0030-BBa_E1010-'''BBa_B1006''' | **BBa_B0030-BBa_E1010-'''BBa_B1006''' | ||
{| | {| | ||
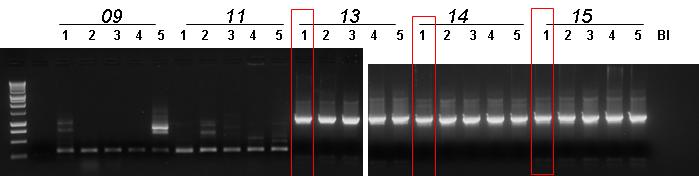
| - | |[[Image:pv_pcr_09_11_13_14_15.jpg|thumb|650px|left|Colony PCR: Marker (1), empty (2) BBa_J23100-'''BBa_B0030'''-BBa_C0012, BBa_J23100-'''BBa_B0030'''-BBa_I15010, '''BBa_B0030'''-BBa_E0040-BBa_B1006, '''BBa_B0030'''-BBa_C0051- | + | |[[Image:pv_pcr_09_11_13_14_15.jpg|thumb|650px|left|Colony PCR: Marker (1), empty (2) BBa_J23100-'''BBa_B0030'''-BBa_C0012, BBa_J23100-'''BBa_B0030'''-BBa_I15010, '''BBa_B0030'''-BBa_E0040-BBa_B1006, '''BBa_B0030'''-BBa_C0051-BBa_B0030, '''BBa_B0030'''-BBa_E1010-BBa_B1006, blank]] |
|} | |} | ||
| Line 61: | Line 61: | ||
**No true positives for BBa_J23100-'''BBa_B0030'''-BBa_I15010 | **No true positives for BBa_J23100-'''BBa_B0030'''-BBa_I15010 | ||
**Non pure true positives for BBa_B0030-BBa_E0040-'''BBa_B1006''' | **Non pure true positives for BBa_B0030-BBa_E0040-'''BBa_B1006''' | ||
| - | **Pure true positives for BBa_B0030-BBa_C0051-''' | + | **Pure true positives for BBa_B0030-BBa_C0051-'''BBa_B0030''' |
**Non pure true positives for BBa_B0030-BBa_E1010-'''BBa_B1006''' | **Non pure true positives for BBa_B0030-BBa_E1010-'''BBa_B1006''' | ||
| Line 77: | Line 77: | ||
|BBa_C0012 | |BBa_C0012 | ||
|BBa_R0062 | |BBa_R0062 | ||
| - | |BBa_B0030-BBa_C0051-''' | + | |BBa_B0030-BBa_C0051-'''BBa_B0030''' |
|BBa_J23100-'''BBa_B0030''' | |BBa_J23100-'''BBa_B0030''' | ||
|} | |} | ||
| Line 97: | Line 97: | ||
|BBa_C0012 | |BBa_C0012 | ||
|BBa_R0062 | |BBa_R0062 | ||
| - | |BBa_B0030-BBa_C0051-''' | + | |BBa_B0030-BBa_C0051-'''BBa_B0030''' |
|BBa_J23100-'''BBa_B0030''' | |BBa_J23100-'''BBa_B0030''' | ||
|} | |} | ||
| Line 112: | Line 112: | ||
|BBa_C0012 (X-P) | |BBa_C0012 (X-P) | ||
|BBa_R0062 (E-X) | |BBa_R0062 (E-X) | ||
| - | |BBa_B0030-BBa_C0051-''' | + | |BBa_B0030-BBa_C0051-'''BBa_B0030''' (S-P) |
|BBa_J23100-'''BBa_B0030''' (E-S) | |BBa_J23100-'''BBa_B0030''' (E-S) | ||
|- | |- | ||
| Line 119: | Line 119: | ||
|BBa_B0030-BBa_C0061-BBa_B1006 (E-S) | |BBa_B0030-BBa_C0061-BBa_B1006 (E-S) | ||
|} | |} | ||
| + | |||
| + | *Gel run/cut. | ||
| + | |||
| + | *Gel extraction for these 11 parts. | ||
| + | |||
| + | *(We already had BBa_I15009(X-P) and BBa_B1006(E-X)) | ||
| + | |||
| + | *Ligations: | ||
| + | #'''BBa_B0030'''-BBa_I15009 | ||
| + | #BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''' | ||
| + | #BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''' | ||
Revision as of 11:05, 20 July 2008
Notebook
| Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 |
|---|---|---|---|---|---|---|
| Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 14 |
Week 8: 07/7/08 - 07/12/08
07/7/08
- Colony PCR (5 colonies for each plate) for:
- BBa_J23100-BBa_B0030-BBa_C0012 (for the second time, we hoped to find true positive colonies)
- BBa_J23100-BBa_B0030-BBa_I15010 (for the second time, we hoped to find true positive colonies)
- BBa_B0030-BBa_E0040-BBa_B1006
- BBa_B0030-BBa_C0051-BBa_B0030
- BBa_B0030-BBa_E1010-BBa_B1006
- Gel results:
- No true positives for BBa_J23100-BBa_B0030-BBa_C0012
- No true positives for BBa_J23100-BBa_B0030-BBa_I15010
- Non pure true positives for BBa_B0030-BBa_E0040-BBa_B1006
- Pure true positives for BBa_B0030-BBa_C0051-BBa_B0030
- Non pure true positives for BBa_B0030-BBa_E1010-BBa_B1006
- We chose to keep the first colony for all the 3 working ligation plates.
- NOTE: BBa_B0030-BBa_E0040-BBa_B1006 and BBa_B0030-BBa_E1010-BBa_B1006 are final parts; we decided to sequence these 2 parts even if gel showed a weak contamination: sequencing results will tell us if there are false positive plasmids in those colonies or if the contamination was only a PCR contamination.
- We infected 9 ml LB + suitable antibiotic with 30 µl of these glycerol stocks:
| BBa_B0030 | BBa_I15010 | BBa_B0030-BBa_E0040-BBa_B1006 (1) | BBa_B0030-BBa_E1010-BBa_B1006 (1) |
| BBa_C0012 | BBa_R0062 | BBa_B0030-BBa_C0051-BBa_B0030 | BBa_J23100-BBa_B0030 |
- Tomorrow we will be ready to perform NINE LIGATIONS...!;)
07/8/08
- We received sequencing results for BBa_B0030-BBa_C0078: sequence showed an extra DNA fragment between BBa_C0078 and Suffix...We decided to ignore it because there was a stop codon before that fragment.
- Glycerol stocks for:
| BBa_B0030 | BBa_I15010 | BBa_B0030-BBa_E0040-BBa_B1006 (1) | BBa_B0030-BBa_E1010-BBa_B1006 (1) |
| BBa_C0012 | BBa_R0062 | BBa_B0030-BBa_C0051-BBa_B0030 | BBa_J23100-BBa_B0030 |
- Miniprep for these parts.
- Plasmid digestion for:
| BBa_B0030 (S-P) | BBa_I15010 (X-P) | BBa_B0030-BBa_E0040-BBa_B1006 (1) (E-X) | BBa_B0030-BBa_E1010-BBa_B1006 (1) (E-X) |
| BBa_C0012 (X-P) | BBa_R0062 (E-X) | BBa_B0030-BBa_C0051-BBa_B0030 (S-P) | BBa_J23100-BBa_B0030 (E-S) |
| BBa_J23100-BBa_B0030-BBa_C0040 (E-S) | BBa_R0051-BBa_B0030-BBa_C0062 (E-S) | BBa_B0030-BBa_C0061-BBa_B1006 (E-S) |
- Gel run/cut.
- Gel extraction for these 11 parts.
- (We already had BBa_I15009(X-P) and BBa_B1006(E-X))
- Ligations:
- BBa_B0030-BBa_I15009
- BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006
- BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006
 "
"