Team:Valencia/Parts
From 2008.igem.org
Contents |
Construction of Valencia Team Biobricks
Preparing inserts
Total DNA was extracted from our yeast strains.
UCP-1 was amplified by PCR using oligonucleotides matching the sequence and bearing the appropriate Biobrick prefix and suffix.
Primers' sequences (EcoRI and PstI sites in bold):
Forward: 5' gAATTCgCggCCgCTTCTAgATggTgAgTTCgACAACTTC 3';
Reverse: 5' TACTAgTAgCggCCgCTgCAgCTATgTggTgCAgTCCACTg 3'
PCR was conducted as follows:
- A first denaturation cycle
- 94º 3'
Followed by 30 amplification cycles:
- 94º 30
60º 1' 30
72º 1'
And a final extension step:
- 72º 10'
Results:
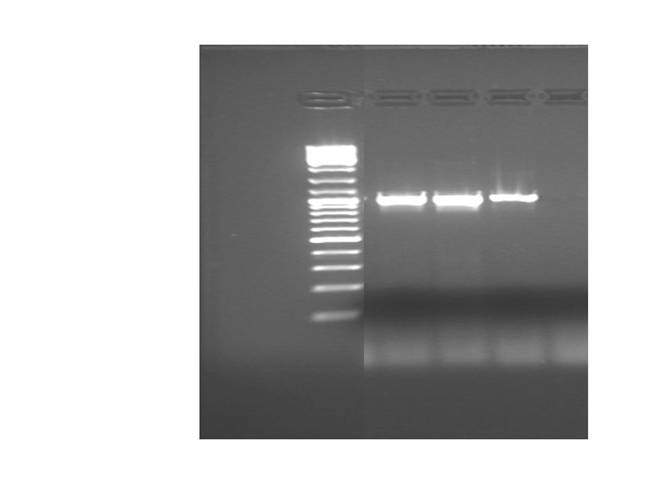
Intense amplicons, of the expected size, (about 1 kb) were produced for UCP-1, 175-deleted and 76 deleted.
Amplicons were digested (H buffer) with EcoRI y PstI.
Preparing vectors
Competent cells were transformed with: pSB1AK3 and pUC18 plasmids
Plasmids were extracted (High pure miniprep plasmid isolation kit ROCHE)
Plasmid were digested with EcoRI and PstI
Ligating Biobricks into plasmids
Both plasmids and inserts were run into 0.8% 0.5X TBE agarose gels and DNA bands excised with a clean scalpel. DNA was extracted from agarose blocks (ultra clean gel spin, DNA purification Kit, MO BIO laboratories).
T4 Ligase was used to ligate inserts and vectors for 1 h at room temperature (2X quick buffer was used).
Competent cells were transformed and resulting colonies (Amp LB) screened with Fw and Rv primers to confirm the presence of inserts.
pSB1AK3 containing UCP-1, 175-deleted and 76-deleted were sent to the Registry
 "
"