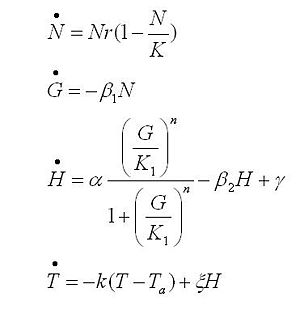Team:Valencia/Parts/UCP76deleted
From 2008.igem.org
| (One intermediate revision not shown) | |||
| Line 8: | Line 8: | ||
| - | ==UCP | + | ==UCP Gly76Δ== |
| Line 85: | Line 85: | ||
| - | ''' | + | '''76 deleted own parameters''' |
Latest revision as of 22:55, 29 October 2008
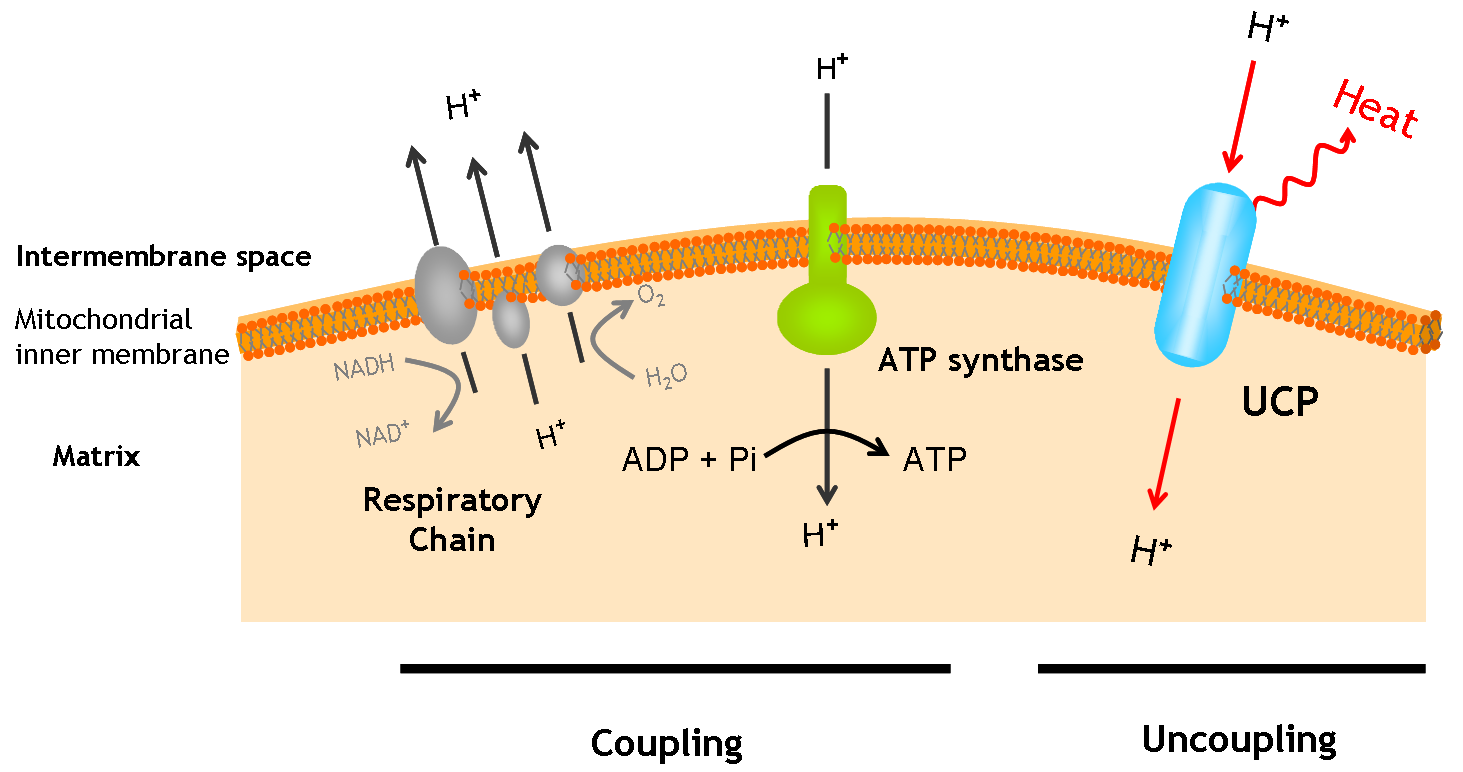
UCP Gly76Δ
The uncoupling protein UCP 76 deleted is a proton carrier which is obtaines from the direct mugenesis of the Ucp1 gene. Ucp 76 has a deletion in the triplet the encodes for the Glycine 76. The mutant shows a generation time and an heat up capacity higher than UCP1. UCP 76 deleted uncouples the respiratory chain of ATP production, converting the metabolic energy in heat. The mutant shows a generation time and an heat up capacity higher than UCP1.
UCP 76 deleted is a 33kd protein.
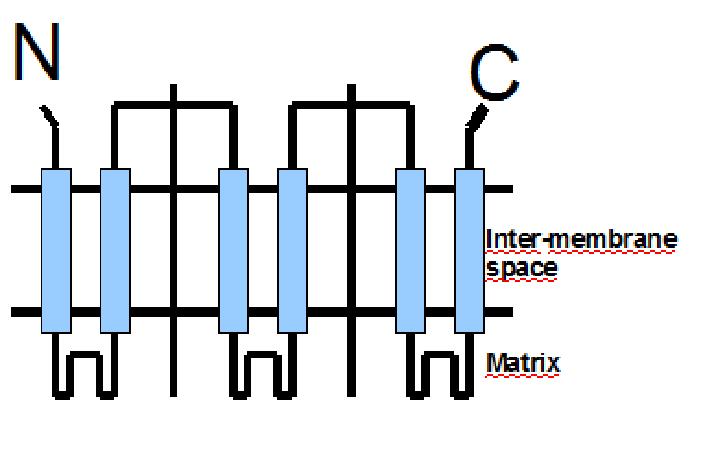
The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.
|
The main difference between UCP 76 deleted and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP 76 deleted proteins.
The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix.
That protein is without direct regulation and its generation time depends on the inner activity the protein.
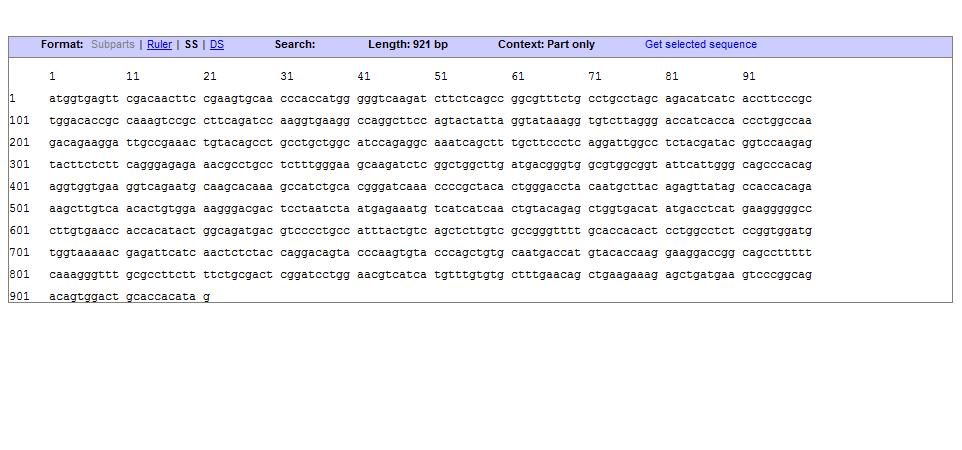
Sequence:
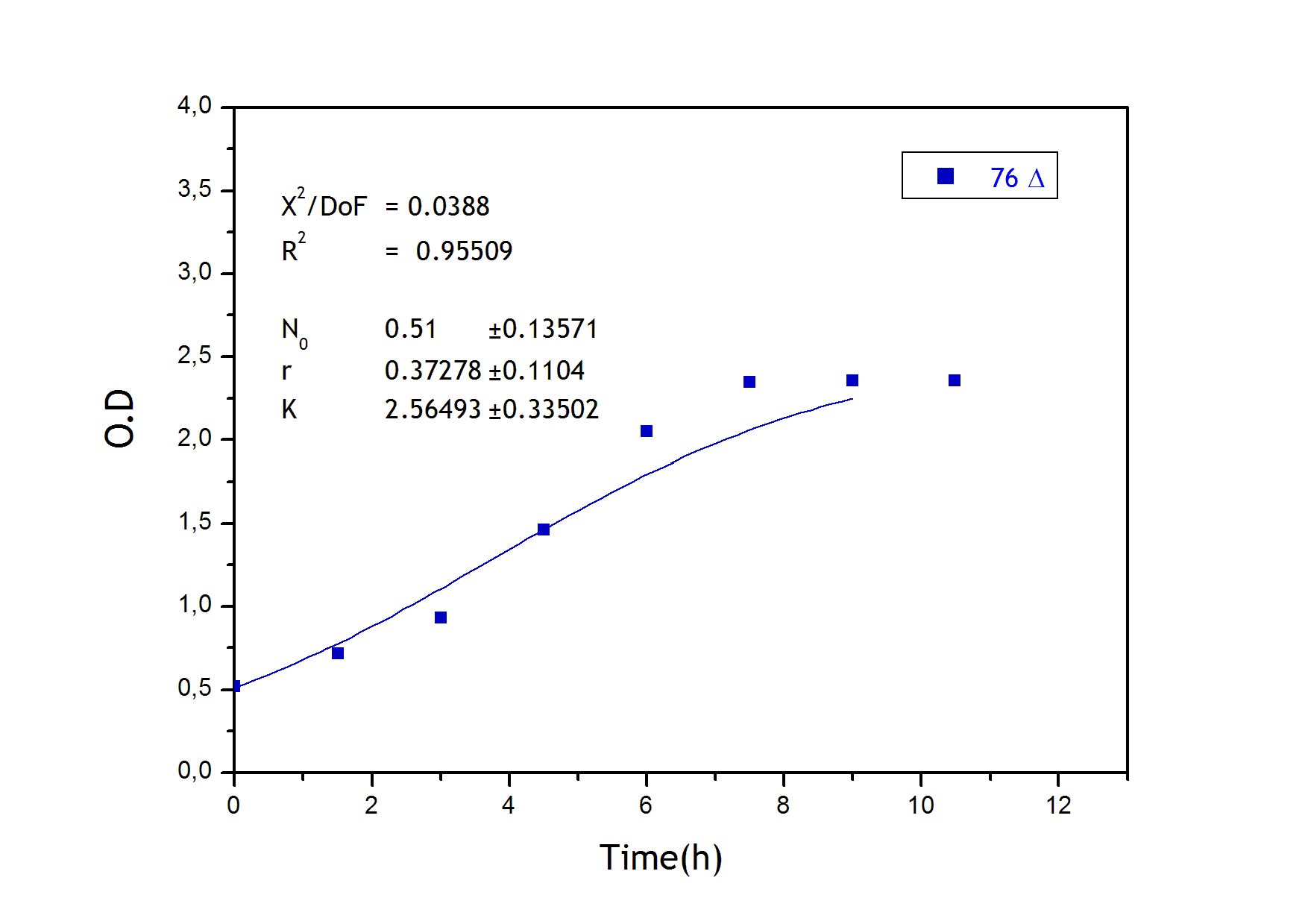
Strains growth
Apart from monitoring temperature evolution, we also characterized O.D. variations of each of our strains. We took O.D. measures every one a half hours for nine hours. We carried out this experiment both in Erlenmeyer flasks in the 30ºC shaking stove and in our LCCs.
Strain growth curve:
Model caonstants and parameters:
The black box model simulates the temperature evolution of the system as a function of the growth rate, galactose concentration and thermogenin expresion. We assume that our system can be reproduced by the following system of equations:
where:
- first equation: Growth of the culture: logistic growth of the different mutants used in our experiments.
- second equation: Galactose evolution: Galactose is the metabolite which induces the thermogenin expresion.
- third equation: Thermogenin concentration level.
- Fourth equation: Temperature evolution. the first term of the equation represents the losses to the ambient of the calorimeter and the second one the temperature increase as a consequence of the thermogenin expresion.
From a sample of 10 experiments in which the evolution of the temperature of the four strain was measured we perform a fit using the software simulink.
Common parameters values:
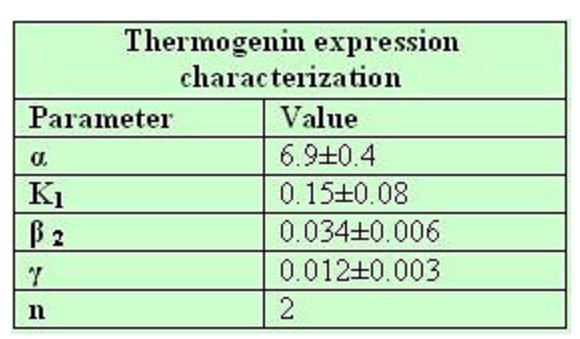
76 deleted own parameters
Galactose comsumption and heat effects.
 "
"