Team:Waterloo/Project
From 2008.igem.org
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Contents |
Overall project
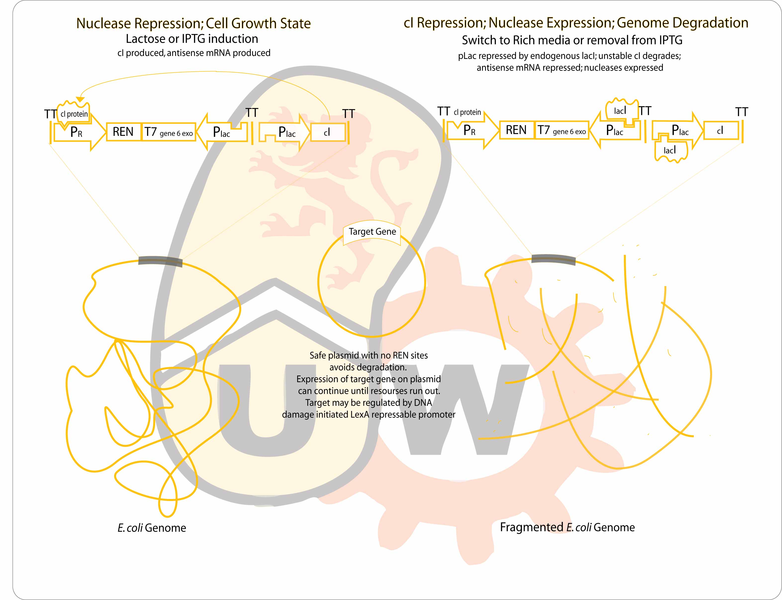
Our goal is to engineer a genome-free, cell-based expression system capable of producing a desired protein in response to environmental signals. The genome will be degraded by the combined activity of a restriction endonuclease (to fragment the genome) and an exonuclease (to hasten degradation of the genome). The gene for the protein of interest will be located on a plasmid which will lack recognition sites for the endonuclease, enabling it to remain intact after genome degradation. Expression of plasmid genes is expected to continue for a period of time until the "cell" expires.
The primary application of this would be an in situ compound production and delivery system for agricultural and/or therapeutic uses.
Construction Strategy
- Already have RFP w/ RBS and TT - should be able to just stick pLac (e .g.) in front of this for preliminary verification. GFP may be a better option for later testing of expression post-kill due to its faster turnover giving better temporal resolution.
- If we did this, we could also use this (in a lacIq background, maybe?) as a preliminary form of control, WT (or lacIq) cell + reporter plasmid (pLac+rfp)
- Test reporter by IPTG induction
- WT (or lacIq) cell + reporter plasmid with nuclease operon (e. g. under TetR control- have some semi-constructed stuff for this, tight enough control for growth?)
- Test for reporter expression upon expression of nuclease (aTe derepression of pTet-controlled nuclease operon)
- Later, could have quorum sensing control T7PoI expression (q.s. + T7 stuff may already exist together in the registry?), and put the nuclease operon under T7 control (instead of TetR)
Tentative stages of construction
Stage 1:
- Obtain gene 6 from T7 or gene D15 from T5 bacteriophage and a suitable REN gene from Psuedomonas mendocina (PmeI) or Psuedomonas alcaligenes (PacI) via PCR with primer-adapters for the biobrick prefix & suffix. T7 may be easier to obtain, and we have been advised that unless we can think of a really good reason to use T5 it isn't worth the trouble of getting T5 over T7.
- To start on the genome degradation side, we can clone the REN (considering PmeI, PacI)and the exonuclease (T5 D15, or T7 gene6)between the lambda pR promoter driving the production of the proper transcript for translation of the genes (which are toxic), and on the other side,pLac (or another lacI repressable promoter) driving production of antisense transcripts to maximize repression of the two nucleases during growth on minimal media with lactose as the sole carbon source. The the unstable cI repressor (BBa_C0051) will be in our construct driven by another instance of a lacI repressable promoter. It should be sufficient to rely on endogenous lacI considering we want induction of pLac at most times, and during production of the nucleases, to destroy the genome, a few antisense transcripts leaking from pLac should not be a problem with the strong lambda promoter making sense transcripts at such a high rate relative to the repressed pLac.
- Transform chassis with the preliminary construct.
- Assay uninduced cell growth. Is our repression strong enough to allow uninduced cells to grow uninhibited?
- We induce for different periods of time (turn off by transferring from lactose to glucose, or glucose to arabinose, depending on system) and assay for genome destruction by a growth test with positive and negative controls (uninduced transformants and no inoculation, respectively); we also take an aliquot, purify the DNA, and run the DNA on a gel alongside the uninduced cells and chassis carrying untransformed plasmid.
- Aside: The tests used to prove sterility by the minicell people for their minicell preparation were plating on growth agar plates, incubating at 37 overnight, and checking for absence of colonies; also inoculating thioglycolate broth with the preparations and incubating for 14 days to demonstrate the absence of any slow-growing organisms (Macdiarmid, et al. 2007)
- Then we have data on how long the nucleases will need to be expressed to destroy the genome. We give the modelling team some work. We get them to help find out exactly where is the optimal spot to integrate our degradation operon into the E. coli genome, based on how long the nuclease genes need to be on, where the REN sites are on the genome, and the rate of degradation by the T5 exonuclease (that data we have).
- We can then subject our safe plasmid with the reporter to the two nucleases in vitro and run on a gel. We could buy the nucleases if we have the budget or maybe we could purify some from our cells that make them, but that might be difficult
Stage 1b (in parallel with stage 1):
- Clone reporter in to plasmid of choice.
- Make sure plasmid has no REN sites for REN in our degradation operon
- Assay plasmid safeness when exposed to nucleases (exonuclease and REN)
Stage 2:
- Construct integrating plasmid with regions of homology, which we had picked based on degradation rates and suitable elements for selection (SacB and antibiotic resistance). Or obtain commercial integrating plasmid to get our stuff in close enough to it's optimal spot.
- Select for cells with integrated genes by plating on media with sucrose and antibiotic
- Assay the viability of uninduced cells again
- Assay genome degradation in much the same way as before, with appropriate controls.
Stage 2b:
- Put reporter under control of LexA repressible promoter or other inducible/repressible promoter system
Stage 3:
- Transform genome degrading chassis (from stage 2)
- Induce genome degradation
- Genome degradation times should be elucidated somewhat, and we should be somewhat able to tell whether or not the reporter is being produced too early (LexA repression). Since both cI and LacI are being used in the degradation regulation, something that will autoregulate would be preferable. There are other options, such as pBAD from that L-ara operon, induced indirectly by arabinose, and not much else.
- The ultimate would be to induce genome degradation, freeze dry, thaw, then induce the plasmid and have the reporter be made (because any practical application of this is going to require a decent shelf life). If the cells were not viable when they left the "manufacturing" facility, it would be preferable to having to induce genome killing in the field.
References:
O'Connor,C.D., & Timmins, K.N. (1987). Highly repressible expression system for cloning genes that specify potentialy toxic proteins. Journal of Bacteriology. 169, 4457-4462.
GUZMAN, L, BELIN, D, CARSON, M. J., & BECKWITH, J (1995). Tight Regulation, Modulation, and High-Level Expression by J. of Bacteriology, 177, 4121-4130.
Macdiarmid, et al. Cancer Cell 11, 431-445, May 2007
 "
"