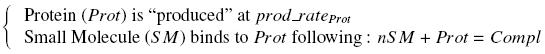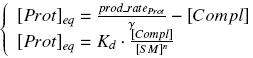Team:Paris/Modeling/Protocol Of Characterization
From 2008.igem.org
|
Protocol of Characterization
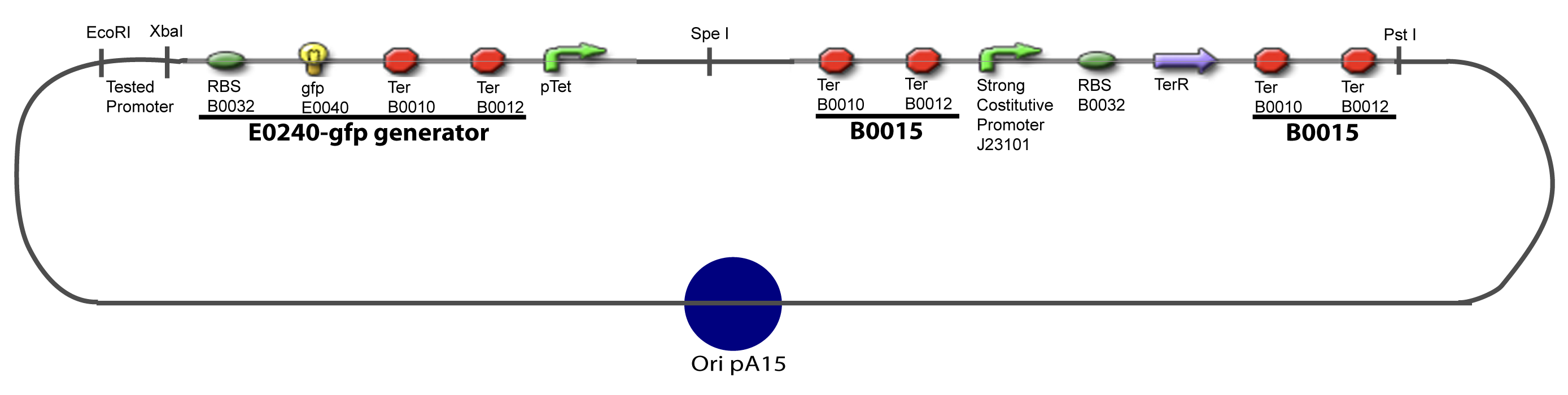
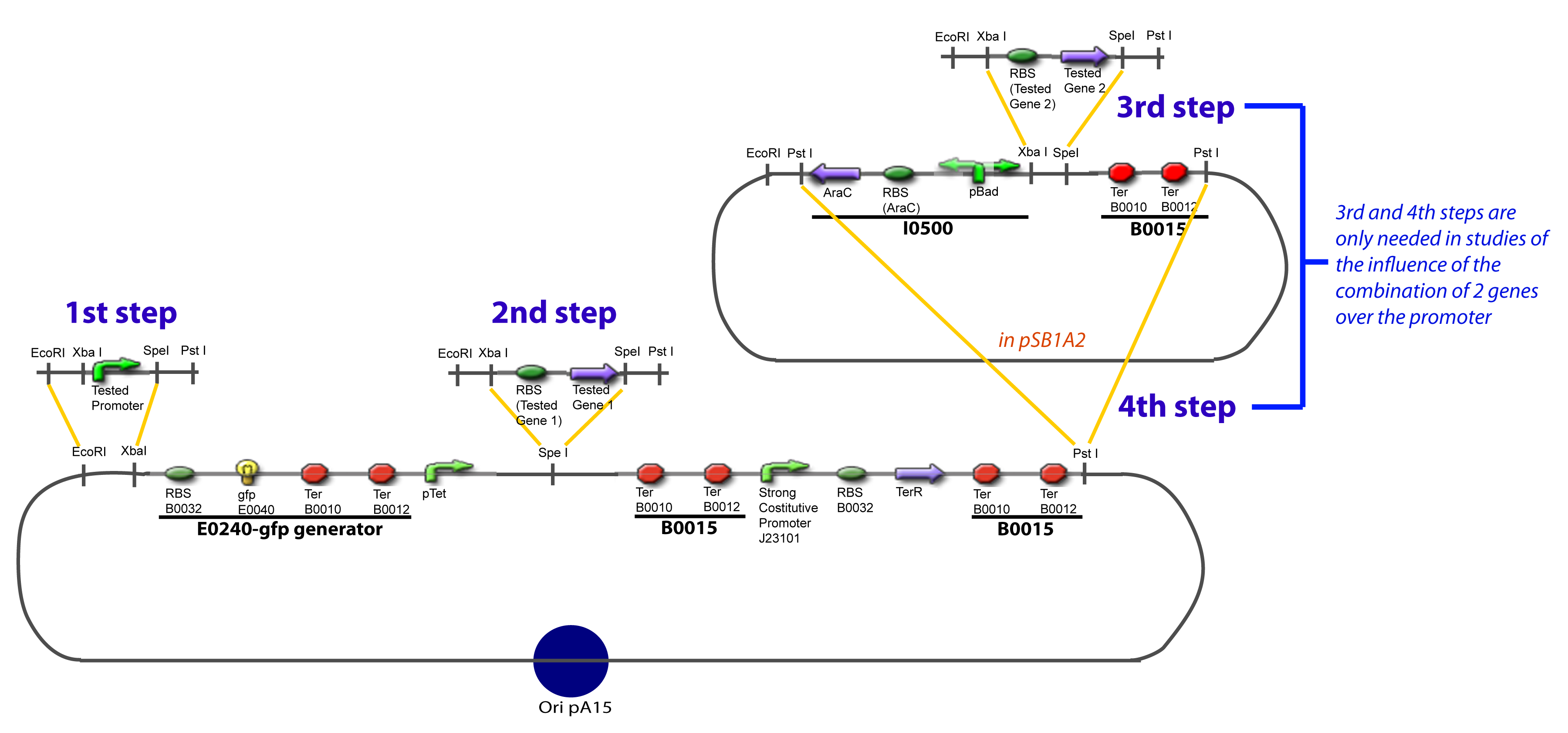
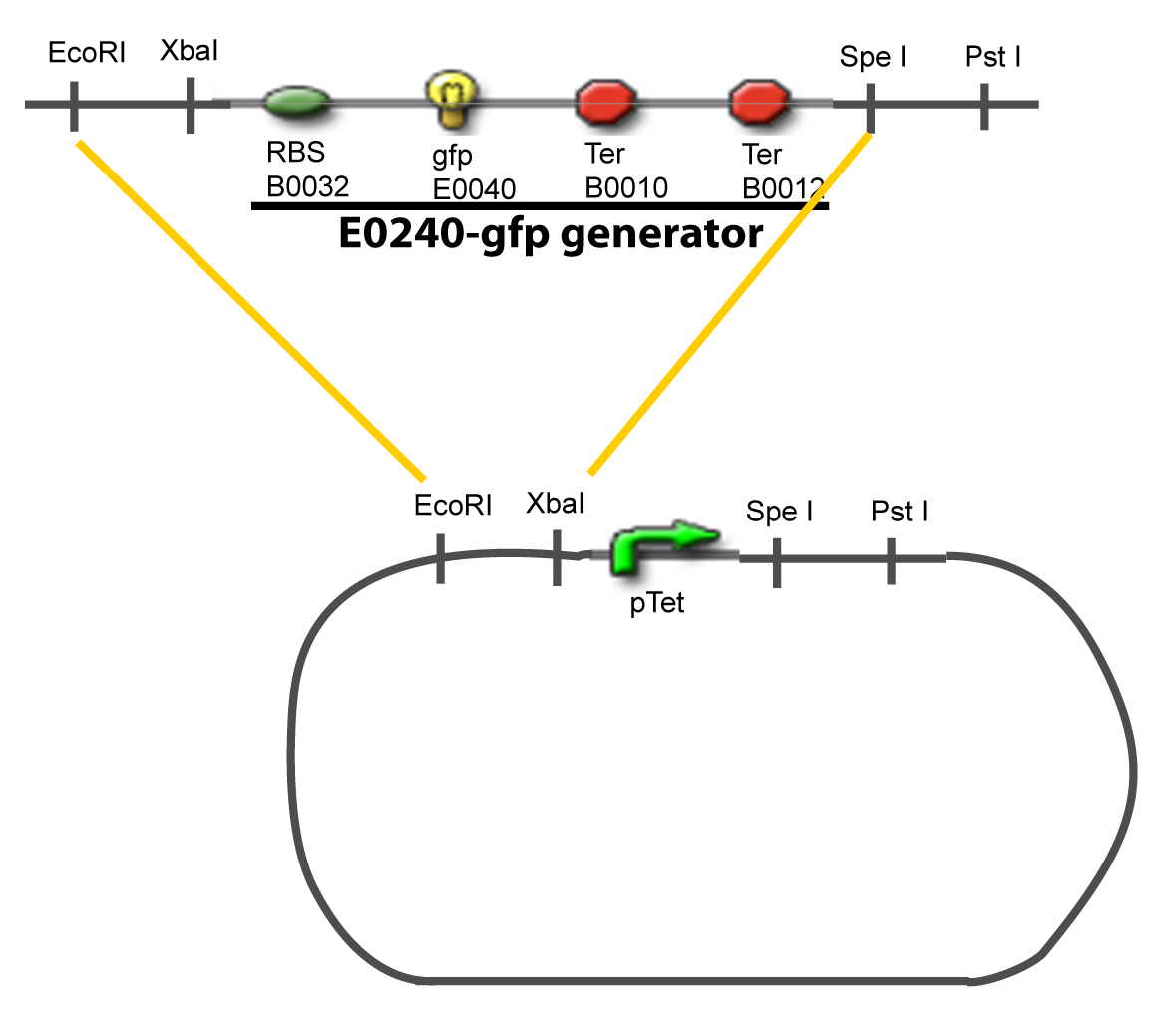
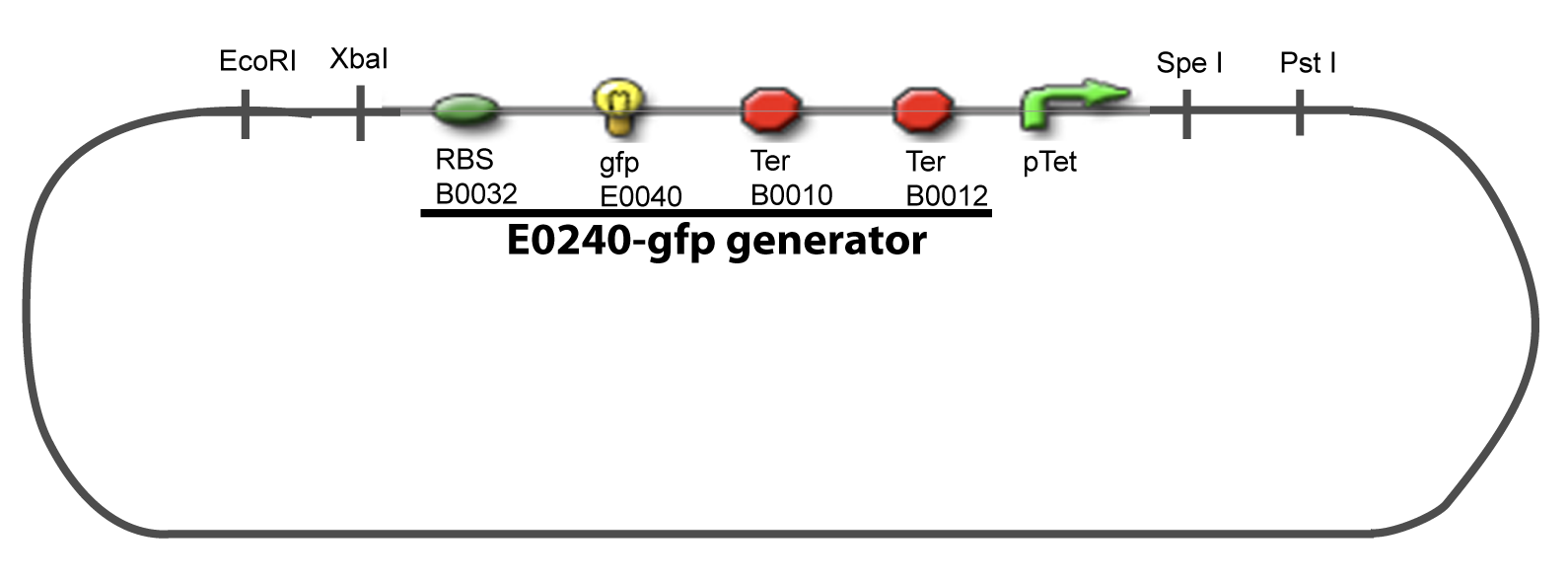
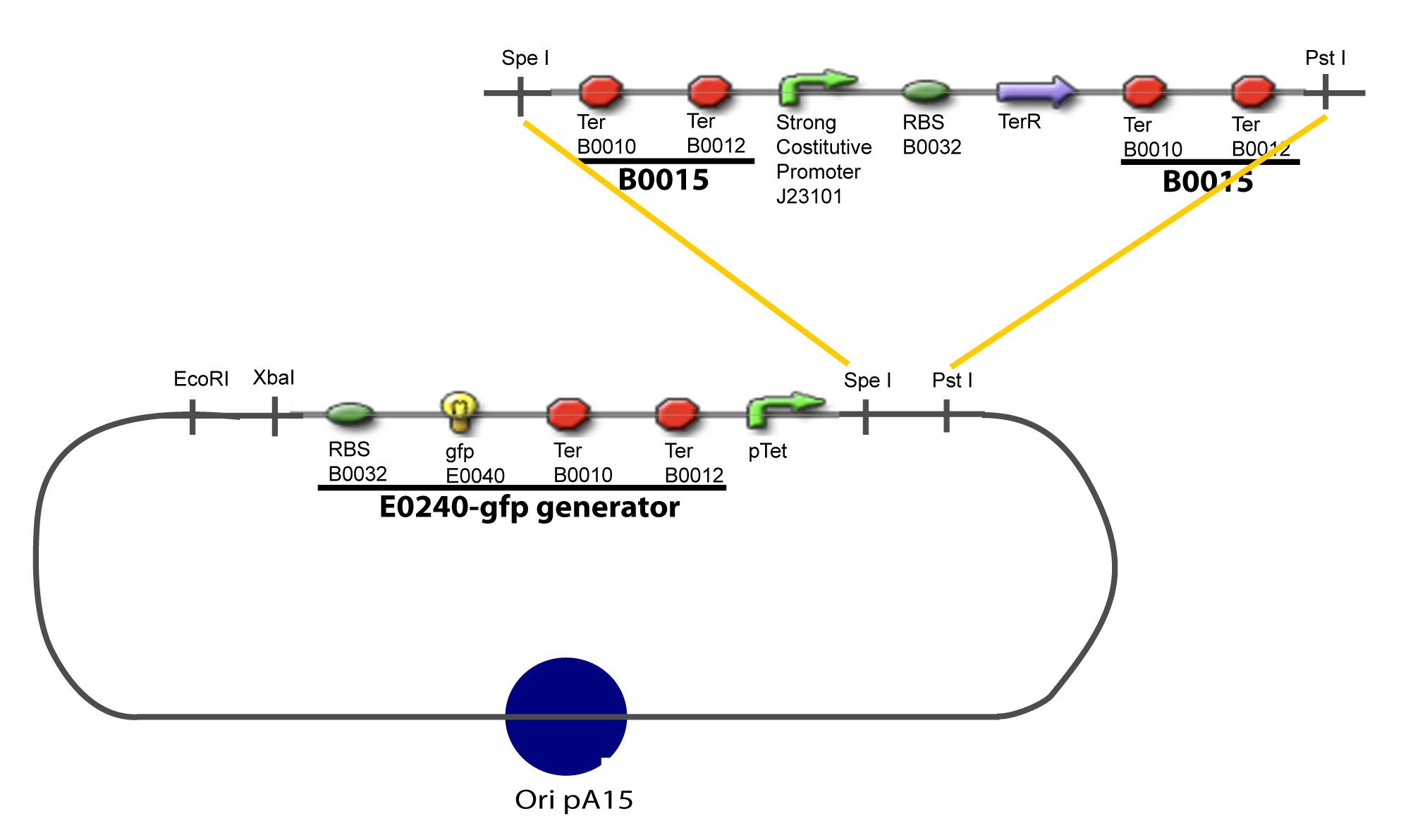
I-Principles of the ExperimentsTo evaluate quantitativly the activity of a promoter in function of its transcription factors, we need data in which the different values of the activities are correlated with various known and controlled values of the transcription factors concentrations. II-Plasmid for promoter characterizationII-A-For study with one Transcription FactorII-B-For study with two Transcription FactorIII-Molecular design for Promoter Characterization PlasmidOur aim is to make this plasmid useful not only for our project but for the whole iGEM comunity. This is why we decided to keep the Biobrick spirit as much as we could, making the plasmid compatible with the parts, so the teams using it needs only the four traditional enzymes: EcoRI, XbaI, SpeI and PstI.We wanted also to make optional the introduction of a second tested transcription factor. The strategy is then based in two plasmids.
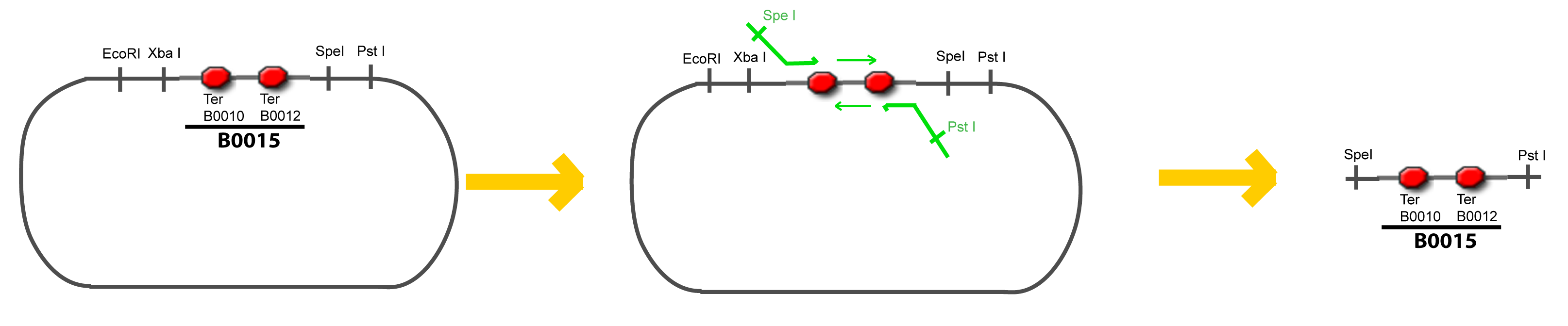
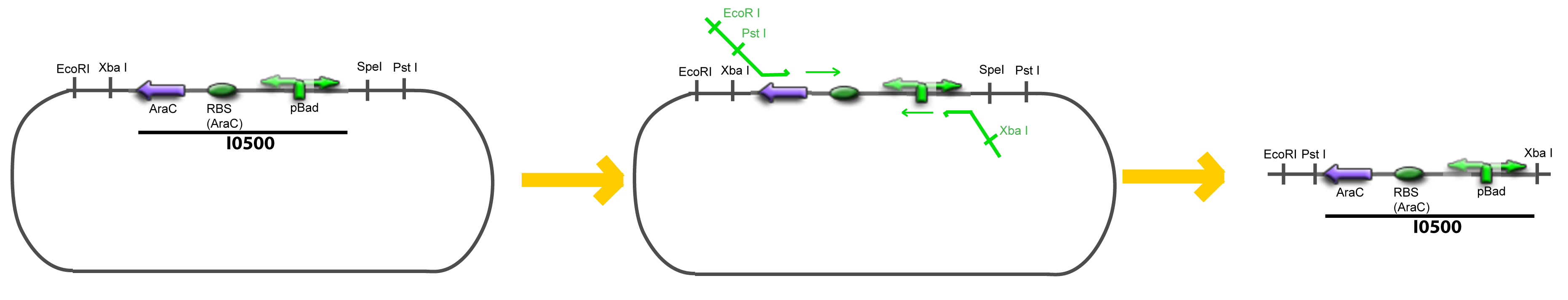
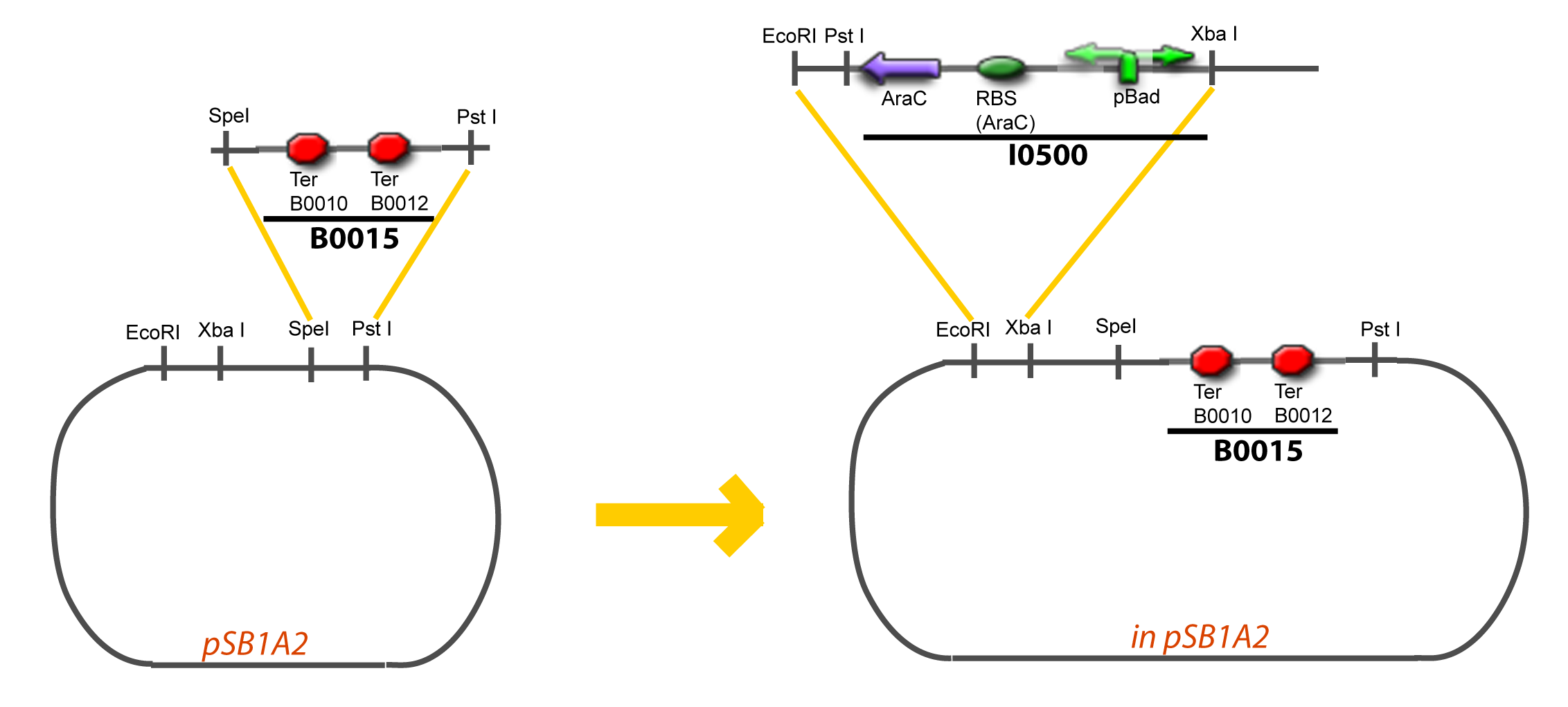
IV-Promoter and Transcription Factors insertionThe strategy to introduce the tested promoter and the tested transcription factor(s) is very simple. The only difficulty is that the order of insertion that we describe has to be respected to avoid unwanted restriction enzyme cuts: ↓ The Steps ↑
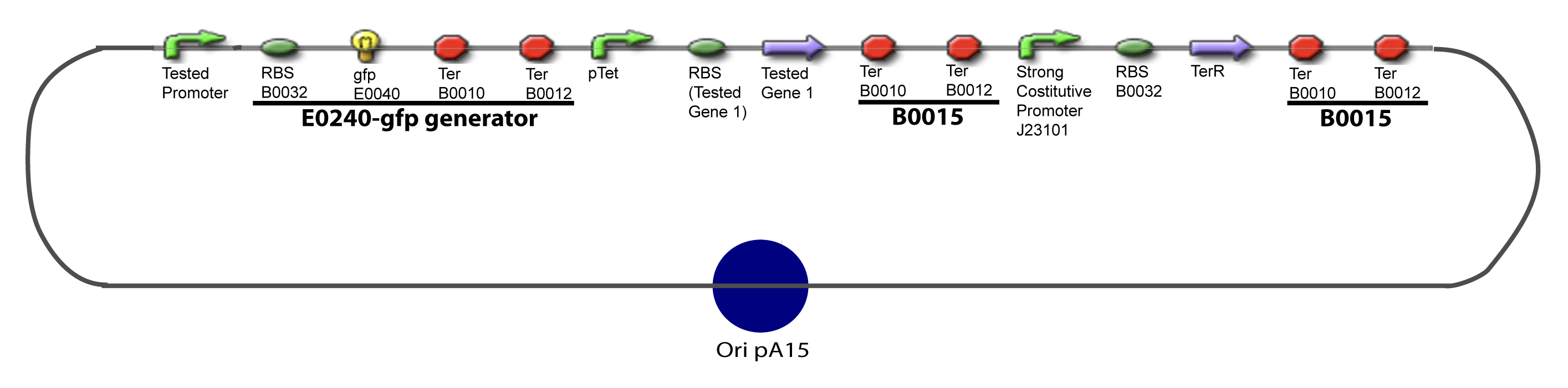
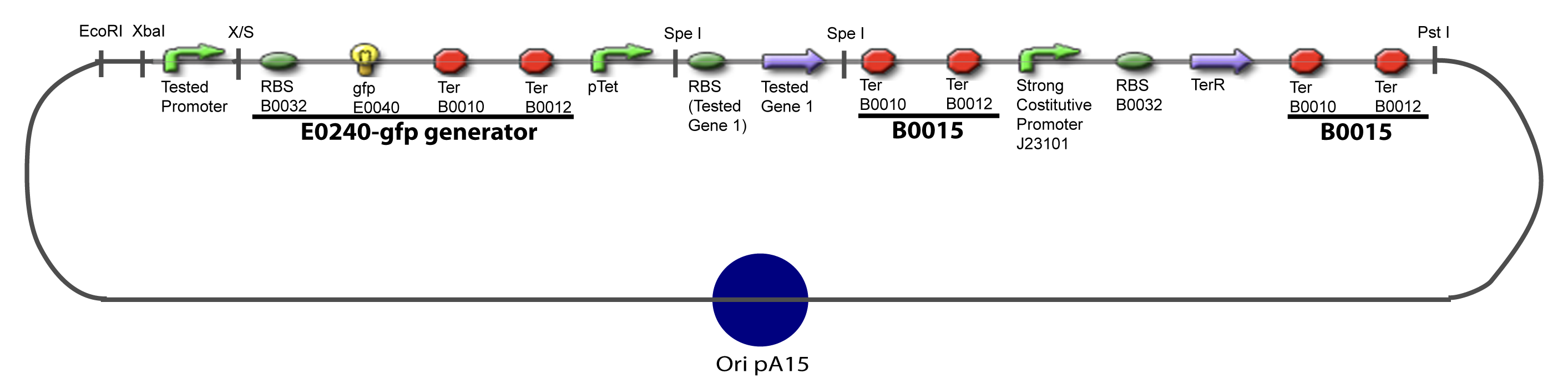
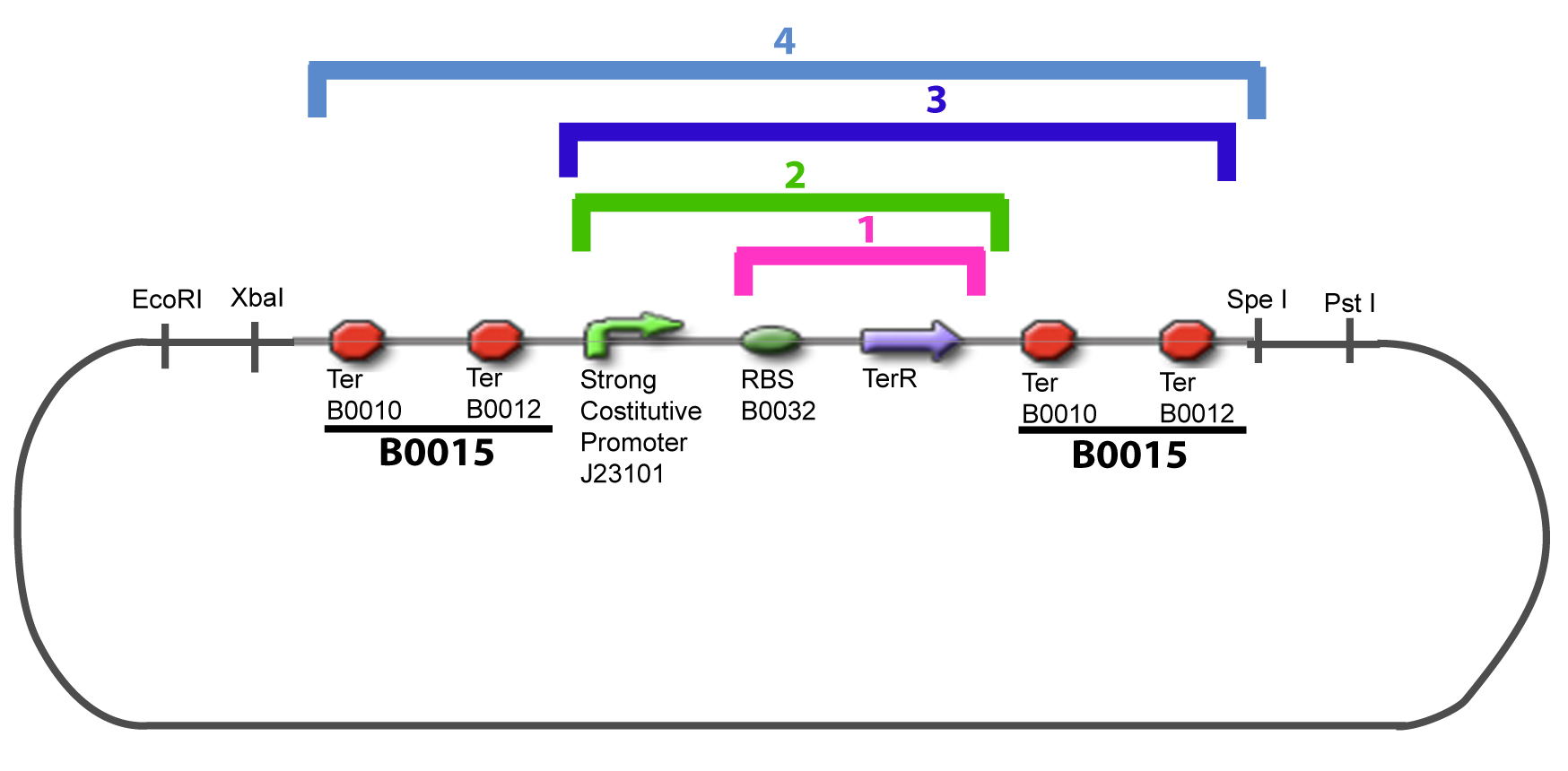
V-Resulting plasmidsV-A-For one transcription factor effect
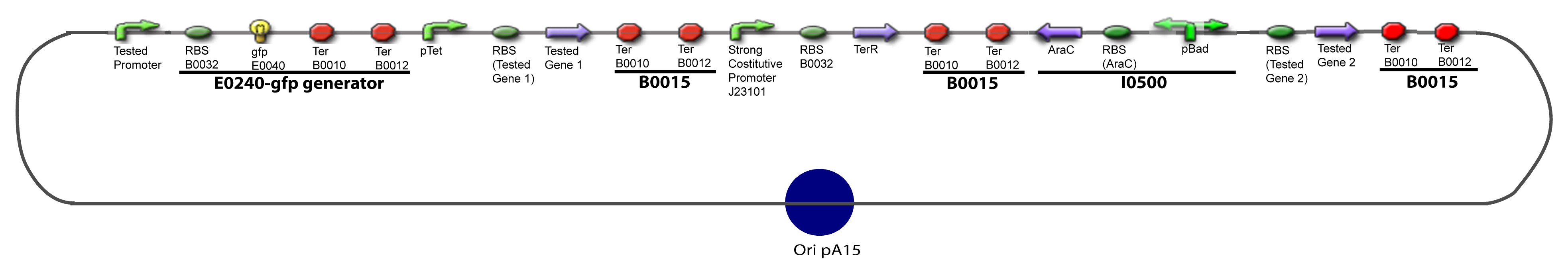
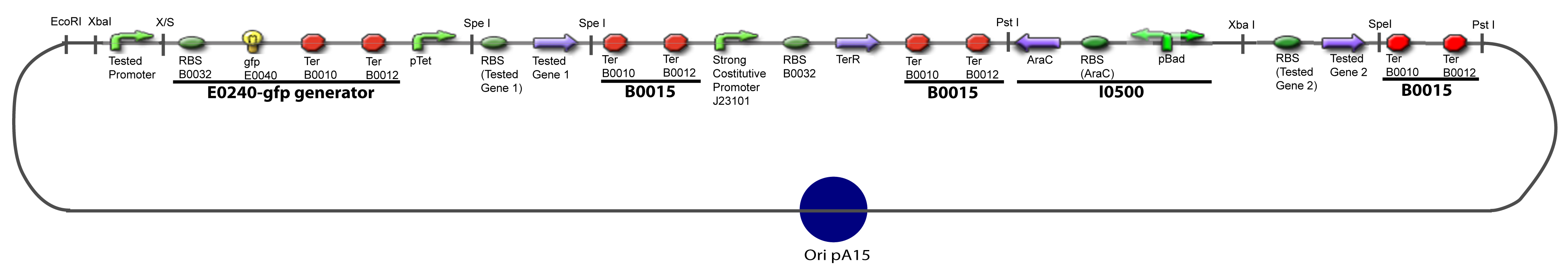
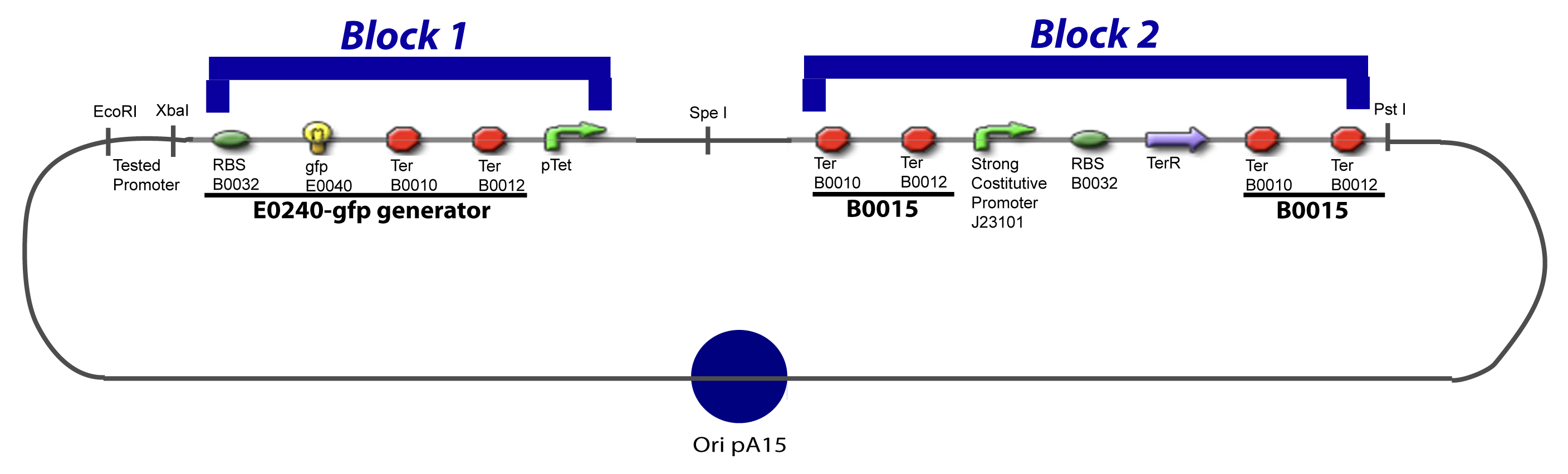
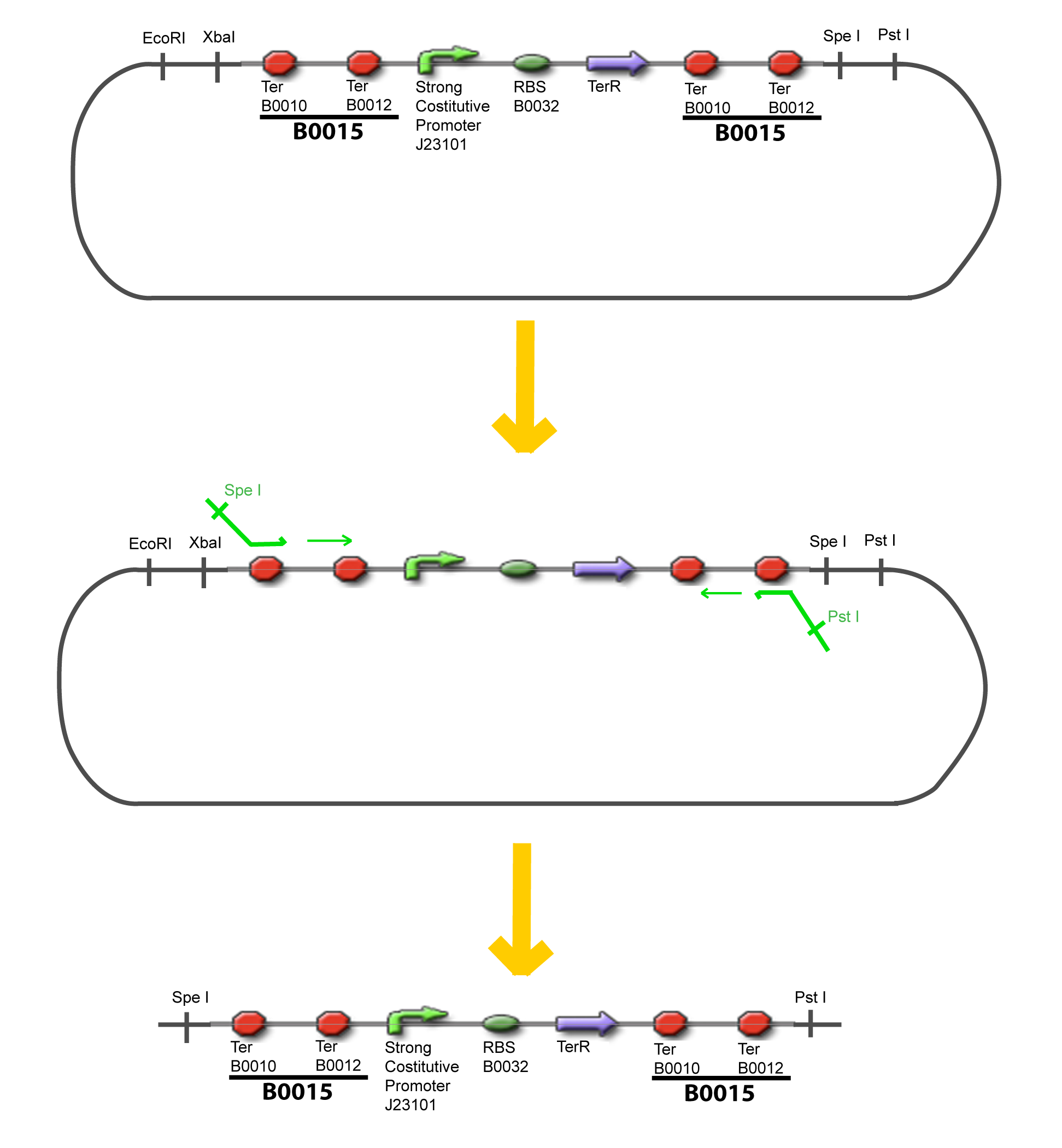
V-B-For two transcription factors effectVI-Protocole for plasmid constructionWe show here our plan to make these plasmids at the experimental level. VI-A-Principal Plasmid constructionWe devided the Principal Plasmid in two main blocks to go faster: VI-A-1-Blocks
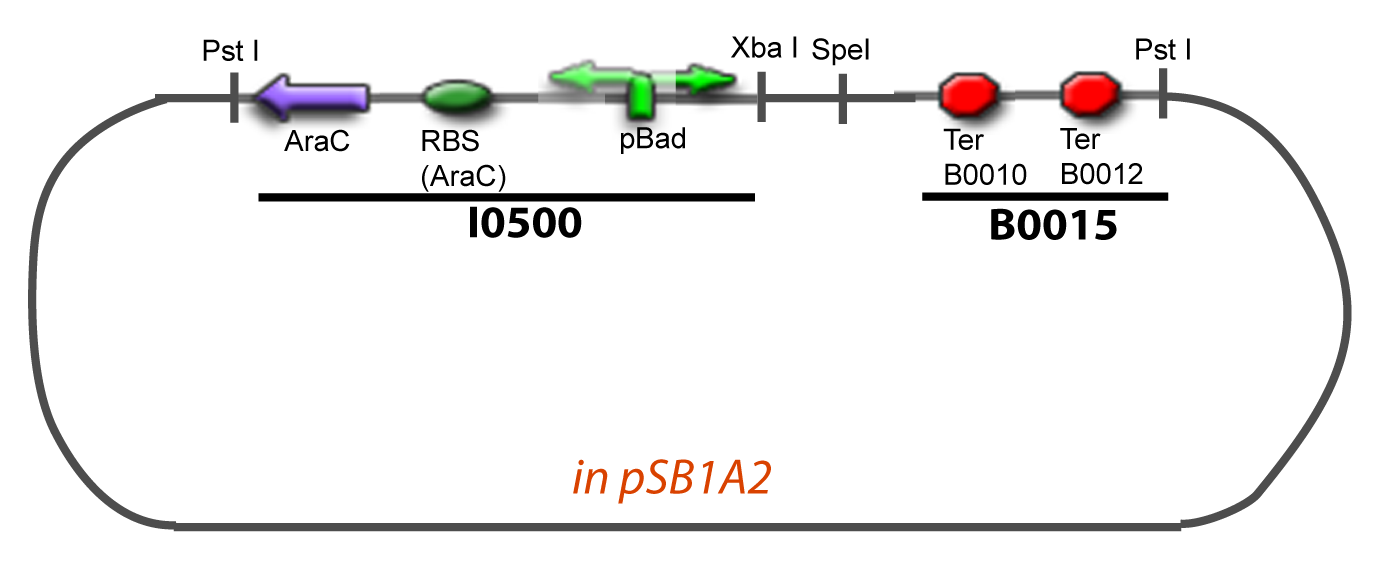
VI-A-4-Two-block assemblyVI-B-Accessory plasmid constructionAddition of restriction sites to B0015 by PCR: Addition of restriction sites to pBAD-AraC by PCR: Assembly: Specific Assumptions for the ExperimentsThe previous experiment is used to find parameters regarding to our modelization. However, the experiments themselves must be described in the same way to interprete these parameters consistently. In this order, the two inductible promoters will be described exactly as the others (those in the system), with similar rules of complexation and transcription. Otherwise, as we control the quantity of transcription factor by the introduction of small molecules, we must model the diffusion/complexation/induction phenomena, which lead from the latter to the former. By putting the cell culture in a chemostat, we will be able to control precisely the concentration of the input small molecules. At quasi steady-state, and assuming a simple, passive and quick diffusion, we consider the inner concentration, the outer medium concentration, and the input concentration beeing all equal. ↓ Quick Mathematical Development ↑
Thanks to these considerations, we can have access to these characterizations : ↓ Prior characterizations ↑
and then ↓ System characterizations ↑
It is important to follow the previous chronological order for the successive characterizations, because some results are reused in the next !
|
 "
"