Team:KULeuven/Model/FullModel
From 2008.igem.org
(→Discussion of the simulation) |
m (→Discussion of the simulation) |
||
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
| + | {{:Team:KULeuven/Tools/Scripting}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| - | <div style=" | + | <div class="floatright" style="margin-top: 110px"><html> |
<object width="250" height="250" > | <object width="250" height="250" > | ||
<param name="movie" value="docbl.swf"> | <param name="movie" value="docbl.swf"> | ||
| Line 21: | Line 23: | ||
<body> | <body> | ||
<p> | <p> | ||
| - | <a href="https://static.igem.org/mediawiki/2008/ | + | <a href="https://static.igem.org/mediawiki/2008/5/5a/FullModelODE.pdf"> |
<img border="0" src="https://2008.igem.org/wiki/skins/common/images/icons/fileicon-pdf.png" width="65" height="60"> | <img border="0" src="https://2008.igem.org/wiki/skins/common/images/icons/fileicon-pdf.png" width="65" height="60"> | ||
</a> | </a> | ||
| Line 104: | Line 106: | ||
<br> | <br> | ||
| - | This simulation can already be seen as a very basic multicell simulation, since it is performed for 1000 cells growing in a microchemostat with a volume of 1000 . For more | + | This simulation can already be seen as a very basic multicell simulation, since it is performed for 1000 cells growing in a microchemostat with a volume of 1000 . For more detailed information, please see our [https://2008.igem.org/Team:KULeuven/Model/MultiCell MultiCell] and [https://2008.igem.org/Team:KULeuven/Model/Diffusion Diffusion] modeling pages. If time is of the essence, the [https://2008.igem.org/Team:KULeuven/Model/Diffusion#Microchemostat Microchemostat subsection] should be most useful. |
==== Discussion of the simulation ==== | ==== Discussion of the simulation ==== | ||
| - | # In the absence of a disease | + | # In the absence of a disease marker, the system starts of with a memory in the OFF state, this means the c2 P22 is produced which shuts down LuxR and ccdB production. LuxI production is also partially eliminated because the memory also outputs antisense LuxI. Put simple, the timer is not active and Dr. Coli can grow without worrying as there is no ccdB and hence no cell death. |
# At t=50.000s the system is activated by a long lightpulse (simulating the insertion of the doctor in the patient's body and sensing desease marker for the first time). This results in: | # At t=50.000s the system is activated by a long lightpulse (simulating the insertion of the doctor in the patient's body and sensing desease marker for the first time). This results in: | ||
## the switching of the memory to the ON state. This means cI 434 instead of c2 P22 so no more antisense LuxI or repression of LuxR and ccdB. (more specific switching simulations can be found in the corresponding [https://2008.igem.org/Team:KULeuven/Model/Memory#Simulations memory] pages). | ## the switching of the memory to the ON state. This means cI 434 instead of c2 P22 so no more antisense LuxI or repression of LuxR and ccdB. (more specific switching simulations can be found in the corresponding [https://2008.igem.org/Team:KULeuven/Model/Memory#Simulations memory] pages). | ||
## With the memory in the ON state, LuxR as well as ccdB will rise to a background level. This level is extremely low for the latter but already significant for the former, because we want to make sure that auto-acivation is possible. | ## With the memory in the ON state, LuxR as well as ccdB will rise to a background level. This level is extremely low for the latter but already significant for the former, because we want to make sure that auto-acivation is possible. | ||
## the activation of the filter (long pulse, the disease marker is strong enough). This is reflected in the production of lactonase, situated just after the filter mechanism. | ## the activation of the filter (long pulse, the disease marker is strong enough). This is reflected in the production of lactonase, situated just after the filter mechanism. | ||
| - | # At t=130.000s the lightpulse is switched | + | # At t=130.000s the lightpulse is switched off (simulating that there's no more disease marker: the patient is cured). This results in: |
## the shutting down of lactonase production, which continues to degrade naturally | ## the shutting down of lactonase production, which continues to degrade naturally | ||
## the shutting down of LacI production, which stops repressing the transcription of LuxI. Slowly, LuxI is produced and thus also 3OC6HSL. | ## the shutting down of LacI production, which stops repressing the transcription of LuxI. Slowly, LuxI is produced and thus also 3OC6HSL. | ||
| Line 119: | Line 121: | ||
# This third part will auto-induce more LuxR receptors and upregulate ccdB. | # This third part will auto-induce more LuxR receptors and upregulate ccdB. | ||
# When the timer has counted to about 50.000s after the input has gone dead, ccdB has reached the lethal amount of around 10-14 molecules per cell and Dr. Coli calls it a day. | # When the timer has counted to about 50.000s after the input has gone dead, ccdB has reached the lethal amount of around 10-14 molecules per cell and Dr. Coli calls it a day. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 13:40, 22 October 2008
Contents |
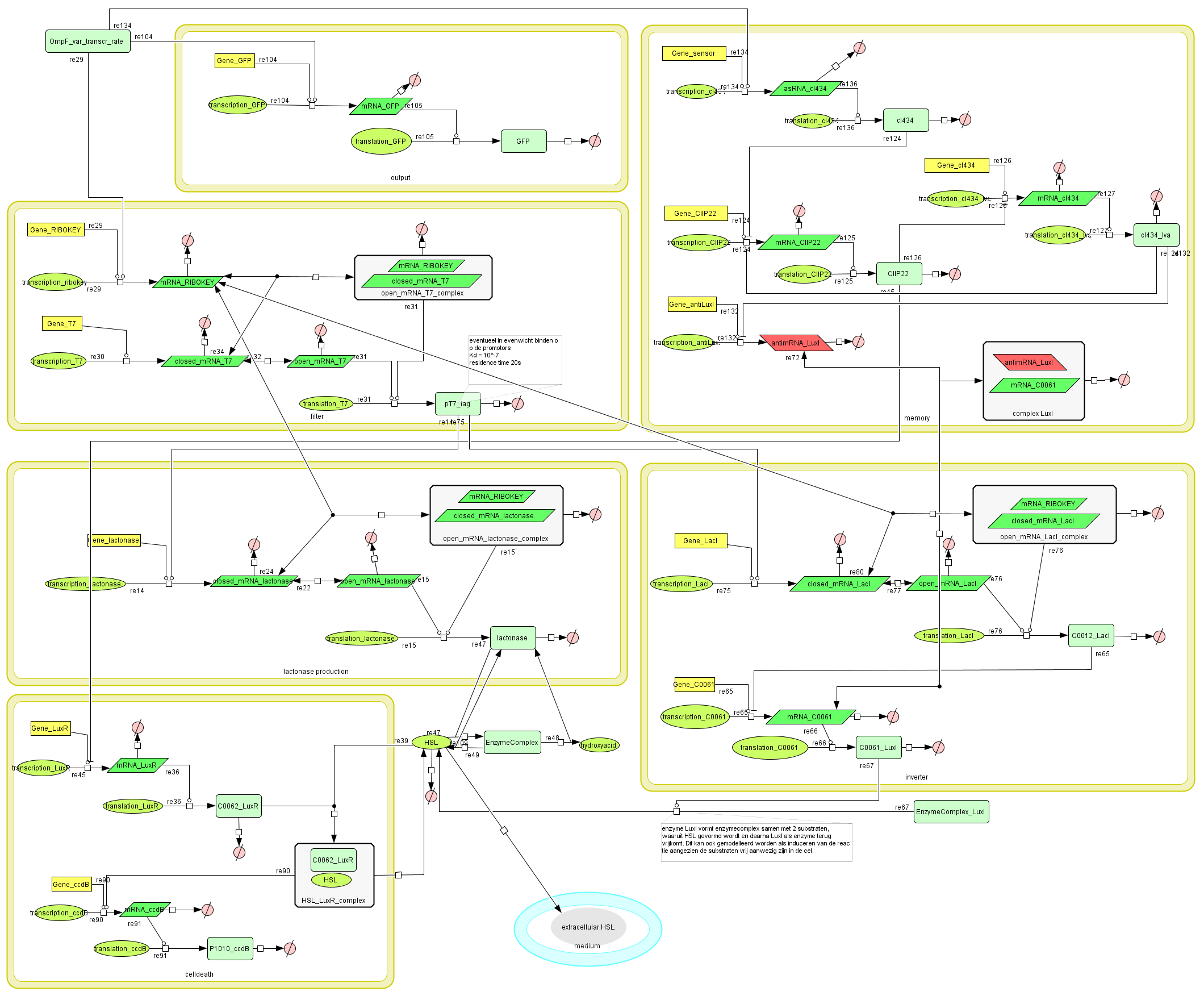
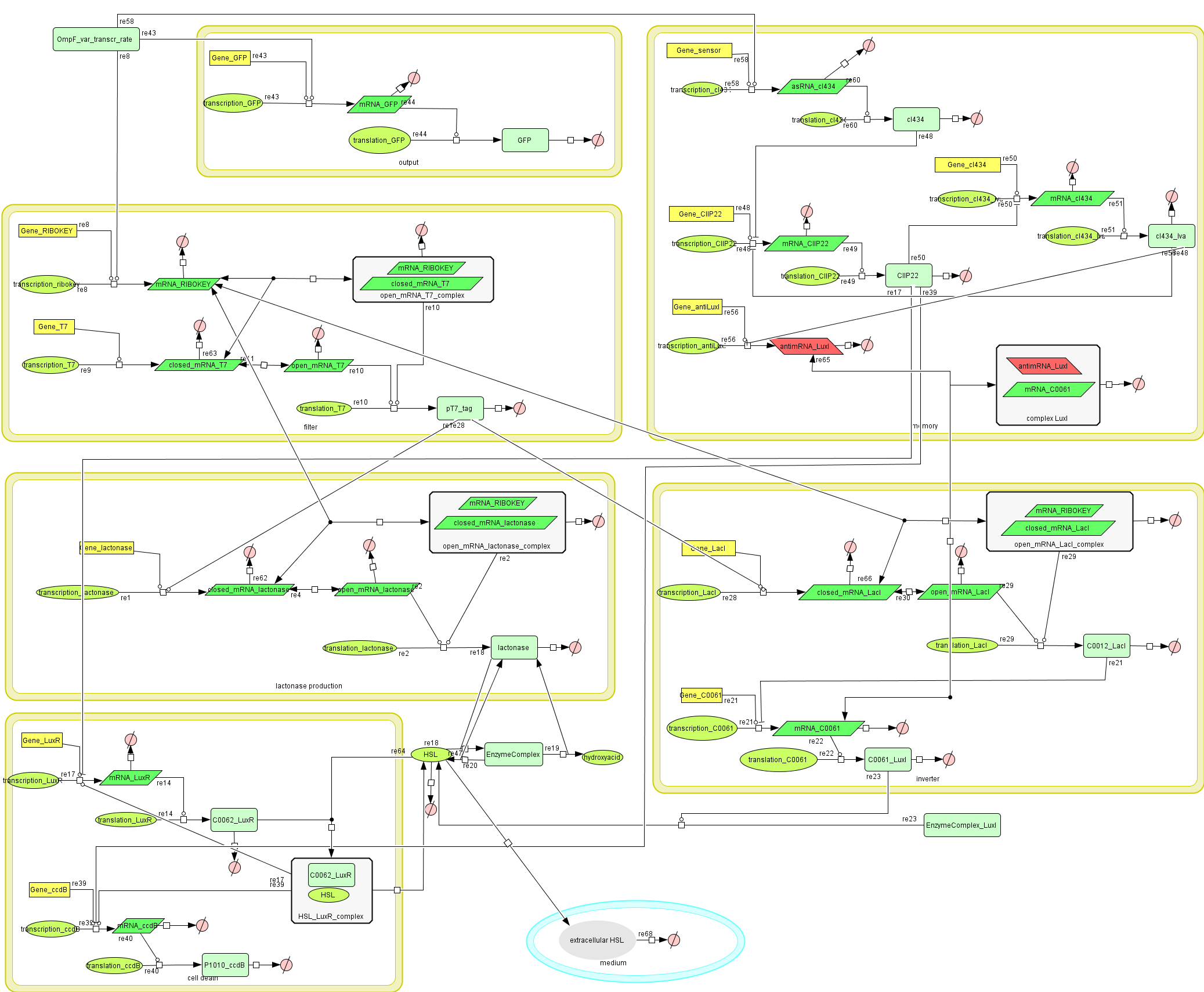
Full Model
Describing the system
Dr. Coli produces a green fluorescent protein (drug) when it senses INPUT (certain disease signal in the human body). When Dr. Coli doesn't sense any INPUT anymore (the patient is cured), the invertimer will function as a molecular timer, counting towards the doctor's own demise: ccdB induced cell death. When INPUT (disease signal) reappers in time, the timer is reset. A filter ensures that the timer is not reset when only “noisy” INPUT signals are sensed. A memory device (stable switch) is included to ensure the clock only starts ticking when it is activated by the first input signal.
ODE's
Parameters
The parameters can be found in the sections about the simple parts of the system.
Models
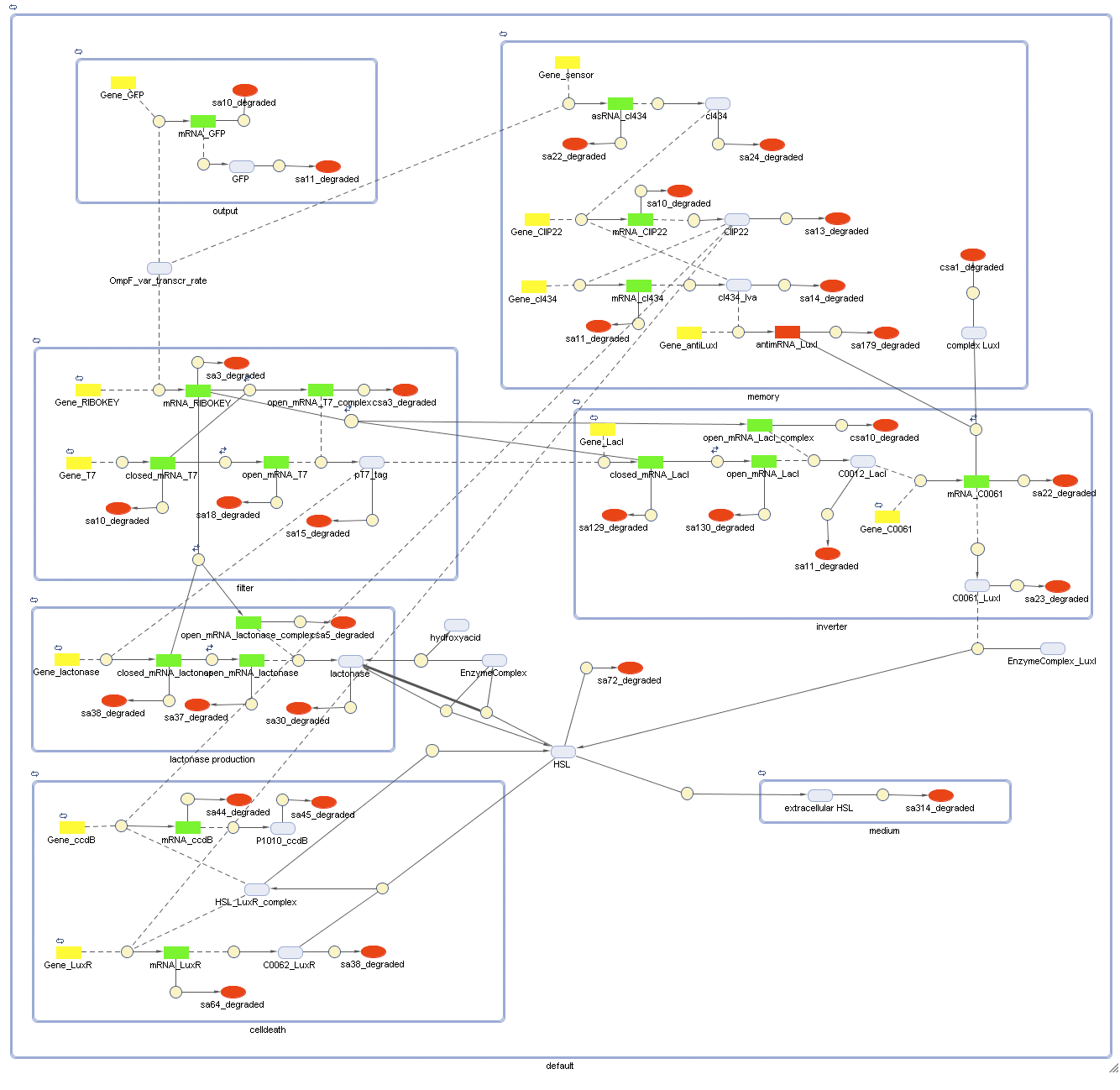
CellDesigner (SBML file)
Matlab (file)
Simulations
Discussion of the simulation:
- In the beginning the memory has status "0" and the simulation clearly shows that the timer isn't activated. This means that the bacteria can grow without any problem (no ccdB-production and hence no cell death) when they haven't sensed any desease marker in the beginning.
- At t=50.000s the system is activated by a long lightpulse (simulating the insertion of the bacteria in the patient's body and sensing desease marker for the first time). This results in:
- switching of the memory to status "1" --> activation of transcription of LuxR (green curve)
- activation of the filter (desease marker is strong enough) which is reflected by the production of lactonase (blue curve), situated just after the filter mechanism.
- At t=75.000 the lightpulse is switched off (simulating that there's no more desease marker: the patient is cured). This results in:
- stopping the production of lactonase, which starts to degrade naturally (blue curve)
- stopping the production of LacI, which stops repressing the transcription of LuxI. In this way HSL can be produced, but, since there's still lactonase present in the cell, there won't be a visible increase in HSL-LuxR-complex because the HSL is being transformed into hydroxyacid.
- At t~=92.000 the lactonase is almost totally degraded which enables the HSL to form a complex with LuxR (yellow curve). This means that we have a timer that starts with a delay of +- 17.000s. At this point the HSL-LuxR-complex really starts to build up gradually (TIMER) for about 10.000s.
- At t~=102.000s the HSL-LuxR-complex peaks, with at the same time (free) LuxR decreasing to almost zero.
- At this point all the newly produced LuxR will go into complex with HSL and no free LuxR will remain. From this point on, the HSL-LuxR-complex will decrease to a steady state value which is equal to the production rate of LuxR-proteins.
- BUT you can see clearly that the ccdB (red curve) also peaks (to a value of 10 to 20 molecules) which is enough to result in cell death.
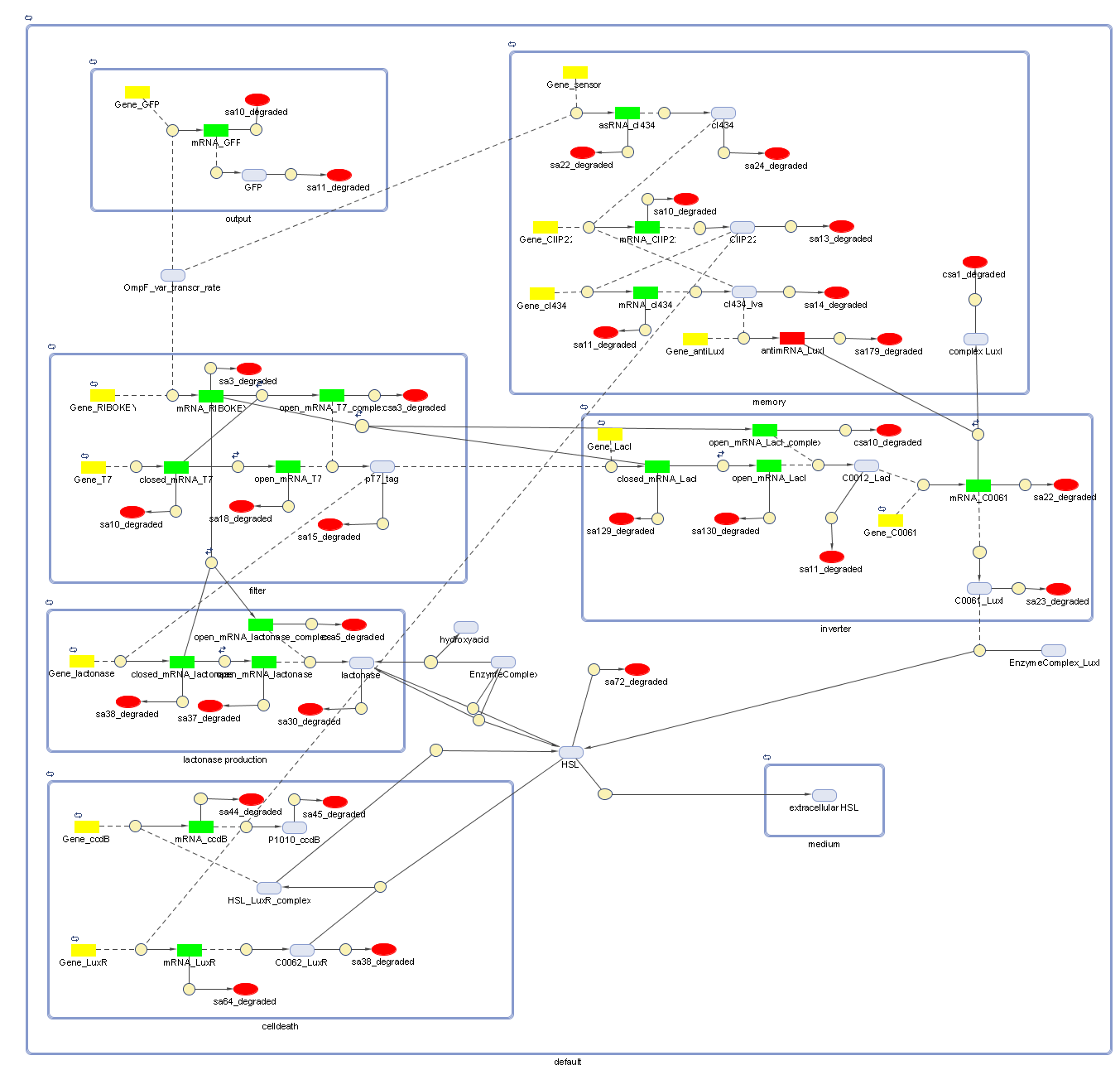
Full Model - Part 2
Extension to the previous model
During the summer we changed a lot of things to our grand scheme. This new system has quite some novelties:
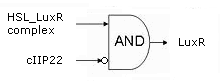
- Cell Death has also gotten an overhaul. Transcription now begins at a new hybrid promoter we made: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K145150 BBa_K145150]. This promoter is repressed by c2 P22, which is produced by the memory in the OFF state, making premature activation and cell death impossible. Besides this repression, the promoter is activated by the HSL-LuxR complex originating from a previously activated timer. The promoter behaves as shown schematically below.
- Second, LuxR is now no longer constitutively produced but is placed behind the beforementioned hybrid promoter. This construction mimicks more closely the natural system where LuxR is upregulated when a threshold amount of HSL is present. Plus it also increases the time it takes to activate ccdB, lengthening the timer. The system will now auto-activate if enough HSL is present and the memory is in the ON state.
- Third, the ccdB coding region is also downstream of the hybrid promoter and is thus also subject to the regulation explained above; c2 P22 repression and HSL-LuxR auto-activation. One difference is that the polymerase must first read through a bad terminator with about 60% efficiency before reaching this coding region. Another difference is in the ribosome binding sites preceding both coding regions. Where LuxR can be translated from a RBS with a relative efficiency of 1.00, the ccdB frame can only be read from a 0.01 efficiency RBS.
Because of this new organisation, we've also turned our attention towards multi cell interactions and diffusion of HSL in the medium again.
Describing the system
ODE's
todo!!!
Parameters
The parameters can be found in the sections about the simple parts of the system.
Extension: parameters of HSL-LuxR auto-activation can be found in Model:New Cell Death.
Models
CellDesigner (SBML file)
Attention: this CellDesigner model is no longer up to date. For the final version, use the MATLAB model below!
Matlab (file)
Simulations



This simulation can already be seen as a very basic multicell simulation, since it is performed for 1000 cells growing in a microchemostat with a volume of 1000 . For more detailed information, please see our MultiCell and Diffusion modeling pages. If time is of the essence, the Microchemostat subsection should be most useful.
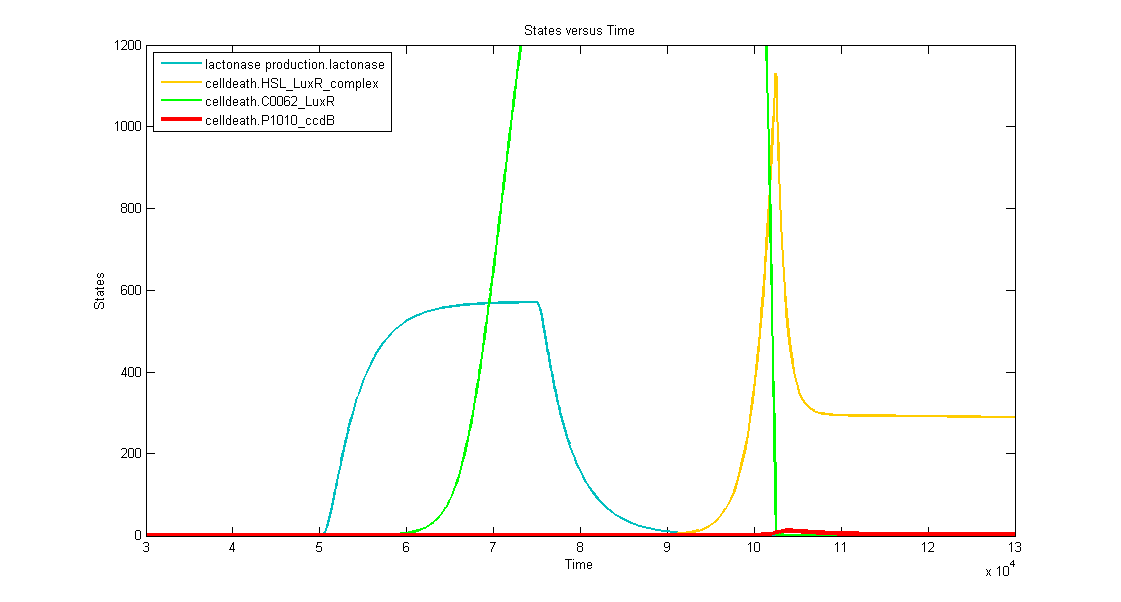
Discussion of the simulation
- In the absence of a disease marker, the system starts of with a memory in the OFF state, this means the c2 P22 is produced which shuts down LuxR and ccdB production. LuxI production is also partially eliminated because the memory also outputs antisense LuxI. Put simple, the timer is not active and Dr. Coli can grow without worrying as there is no ccdB and hence no cell death.
- At t=50.000s the system is activated by a long lightpulse (simulating the insertion of the doctor in the patient's body and sensing desease marker for the first time). This results in:
- the switching of the memory to the ON state. This means cI 434 instead of c2 P22 so no more antisense LuxI or repression of LuxR and ccdB. (more specific switching simulations can be found in the corresponding memory pages).
- With the memory in the ON state, LuxR as well as ccdB will rise to a background level. This level is extremely low for the latter but already significant for the former, because we want to make sure that auto-acivation is possible.
- the activation of the filter (long pulse, the disease marker is strong enough). This is reflected in the production of lactonase, situated just after the filter mechanism.
- At t=130.000s the lightpulse is switched off (simulating that there's no more disease marker: the patient is cured). This results in:
- the shutting down of lactonase production, which continues to degrade naturally
- the shutting down of LacI production, which stops repressing the transcription of LuxI. Slowly, LuxI is produced and thus also 3OC6HSL.
- At t~=150.000, all the lactonase and the LacI are gone and it is possible to see LuxI and HSL increase. This means that the actual 'counting' only starts about 17.000s after the input is switched off. Part of the HSL diffuses outside of the cell, another part remains free but the third part associates with the background of Lux receptors (free LuxR drops).
- This third part will auto-induce more LuxR receptors and upregulate ccdB.
- When the timer has counted to about 50.000s after the input has gone dead, ccdB has reached the lethal amount of around 10-14 molecules per cell and Dr. Coli calls it a day.
 "
"