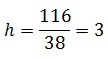Team:Bologna/Wetlab
From 2008.igem.org
Rita.morini (Talk | contribs) (→Introduction) |
|||
| (140 intermediate revisions not shown) | |||
| Line 27: | Line 27: | ||
=Introduction= | =Introduction= | ||
[[Image:Collage2.jpg|700px|right]] | [[Image:Collage2.jpg|700px|right]] | ||
| - | To obtain regulated promoters we synthesized four libraries of operator sequences, respectively for LacI, TetR, cI and LexA repressor proteins. Here is how we designed the operator libraries | + | To obtain regulated promoters we synthesized four libraries of operator sequences, respectively for LacI, TetR, cI and LexA repressor proteins. Here is explained how we designed the operator libraries and isolated single operators to clone them in the standard format. Moreover, we built and tested: a LacI regulated promoter to tune promoter activation and a lexA based promoter to study the SOS activation (UV and peroxidase). |
| - | |||
| - | =Operator | + | |
| + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
| + | |||
| + | =At last... Operator sites as BioBricks!= | ||
Even though transcriptional regulation still plays a pivotal role in synthetic biology, a modular assembly of regulated promoters and the characterization of their properties has not been formalized, yet. At present state, each promoter in the Registry, though complex, is treated as a “standalone” monolithic element. Thus, the choice of a regulated promoter implies a prefixed sigmoid regulation curve. The only option, using the current BioBricks, is to choose a discretional scaling factor in the transfer of the transcription function to the protein through an appropriate RBS. Critically, the choice of one specific transcription factor limits the choice to just one possible promoter. The assembly of regulated promoters as the combination of modular parts, i. e. unregulated promoters and operators, could permit the rapid design of regulatory elements with fixed characteristics. In fact, promoter transcriptional strength and repressor binding affinity could be independently chosen. | Even though transcriptional regulation still plays a pivotal role in synthetic biology, a modular assembly of regulated promoters and the characterization of their properties has not been formalized, yet. At present state, each promoter in the Registry, though complex, is treated as a “standalone” monolithic element. Thus, the choice of a regulated promoter implies a prefixed sigmoid regulation curve. The only option, using the current BioBricks, is to choose a discretional scaling factor in the transfer of the transcription function to the protein through an appropriate RBS. Critically, the choice of one specific transcription factor limits the choice to just one possible promoter. The assembly of regulated promoters as the combination of modular parts, i. e. unregulated promoters and operators, could permit the rapid design of regulatory elements with fixed characteristics. In fact, promoter transcriptional strength and repressor binding affinity could be independently chosen. | ||
| - | A first step in the rationalization of promoter design was done in the iGEM 2007 with the introduction in the Registry of a family of constitutive promoters. Each family element differs from the others just for few base pairs in -35 and -10 regions, keeping constant the rest of the sequence and giving rise to a different level of transcription strength. We decided to use this valuable work as a platform for a deeper and more general promoter design strategy. To obtain regulated promoters we decided to combine these parts with short regulatory sequences that operate as a binding site for the transcription factor. To this aim, we synthesized four libraries of operator sequences, respectively for [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079045 LacI], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079046 TetR], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079047 cI] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079048 LexA] repressor proteins. In each library, there are three sequences (see Table below) each with a different repressor binding affinity to the repressor protein. | + | A first step in the rationalization of promoter design was done in the iGEM 2007 with the introduction in the Registry of a family of constitutive promoters. Each family element differs from the others just for few base pairs in -35 and -10 regions, keeping constant the rest of the sequence and giving rise to a different level of transcription strength. We decided to use this valuable work as a platform for a deeper and more general promoter design strategy. To obtain regulated promoters we decided to combine these parts with short regulatory sequences that operate as a binding site for the transcription factor. To this aim, we synthesized four libraries of operator sequences, respectively for [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079045 LacI], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079046 TetR], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079047 cI] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079048 LexA] repressor proteins ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K079045 see details]). In each library, there are three sequences (see Table below) each with a different repressor binding affinity to the repressor protein. Since the libraries were synthetized on pGA18 and pMA Geneart vectors, we isolated each operator with the intention to clone them into BioBrick standard assembly plasmids. The choice of the operator should give a relative fine-tuning of promoter sensitivity to the repressor. We decided to take the Berkley's costitutive promoter library as a good "collection" from which we could select the unregulated promoter, according with the chosen transcriptional strength. |
<br> | <br> | ||
| Line 42: | Line 44: | ||
|colspan=3| <font size="+1">LacI</font> | |colspan=3| <font size="+1">LacI</font> | ||
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |width= | + | |width=100| '''Name''' |
| - | |width= | + | |width=180| '''Sequence''' |
| - | |width= | + | |width=150| '''Affinity''' |
|- style="background:#f6efcd; color:black" | |- style="background:#f6efcd; color:black" | ||
| - | | [http://partsregistry.org/Part: | + | | [http://partsregistry.org/Part:BBa_K079017 Lac SymL] |
| - | | | + | | aattgtgagcgctcacaatt |
| - | | | + | | Very Strong |
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079018 Lac O1] | ||
| + | | aattgtgagcggataacaatt | ||
| + | | Intermediate | ||
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079019 Lac O2] | ||
| + | | aaatgtgagcgagtaacaacc | ||
| + | | Weak | ||
|} | |} | ||
{| align="right" | {| align="right" | ||
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |colspan=3| | + | |colspan=3| <font size="+1">Tet</font> |
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |width= | + | |width=100| '''Name''' |
| - | |width= | + | |width=180| '''Sequence''' |
| - | |width= | + | |width=150| '''Affinity''' |
|- style="background:#f6efcd; color:black" | |- style="background:#f6efcd; color:black" | ||
| - | | [http://partsregistry.org/Part: | + | | [http://partsregistry.org/Part:BBa_K079036 TeT O] |
| - | | | + | | cctatcagtgataga |
| - | | | + | | Strong |
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079037 TetO-4C] | ||
| + | | cctgtcagtgacaga | ||
| + | | Intermediate | ||
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079038 TetO-wt/4C5G] | ||
| + | | cctatcagtgacgga | ||
| + | | Weak | ||
|} | |} | ||
| - | <br><br><br><br><br><br> | + | <br><br><br><br><br><br><br><br><br> |
{| align="left" | {| align="left" | ||
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |colspan=3| <font size="+1"> | + | |colspan=3| <font size="+1">Lambda</font> |
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |width= | + | |width=100| '''Name''' |
| - | |width= | + | |width=180| '''Sequence''' |
| - | |width= | + | |width=150| '''Affinity''' |
|- style="background:#f6efcd; color:black" | |- style="background:#f6efcd; color:black" | ||
| - | | [http://partsregistry.org/Part: | + | | [http://partsregistry.org/Part:BBa_K079041 Lambda OR1] |
| - | | | + | | tatcaccgccagaggta |
| - | | | + | | Strong/Intermediate cI/Cro |
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079042 Lambda OR2] | ||
| + | | taacaccgtgcgtgttg | ||
| + | | Intermediate/Weak cI/Cro | ||
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079043 Lambda OR3] | ||
| + | | tatcaccgcaagggata | ||
| + | | Intermediate/Intermediate cI/Cro | ||
|} | |} | ||
{| align="right" | {| align="right" | ||
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |colspan=3| | + | |colspan=3| <font size="+1">Lex</font> |
|- style="background: #c09a6d; text-align: center;" | |- style="background: #c09a6d; text-align: center;" | ||
| - | |width= | + | |width=100| '''Name''' |
| - | |width= | + | |width=180| '''Sequence''' |
| - | |width= | + | |width=150| '''Affinity''' |
|- style="background:#f6efcd; color:black" | |- style="background:#f6efcd; color:black" | ||
| [http://partsregistry.org/Part:BBa_K079039 LexA 1] | | [http://partsregistry.org/Part:BBa_K079039 LexA 1] | ||
| - | | | + | | ctgtatatatatacag |
| - | | | + | | Very Strong |
| + | |- style="background:#f6efcd; color:black" | ||
| + | | [http://partsregistry.org/Part:BBa_K079040 LexA 2] | ||
| + | | ctgtatgagcatacag | ||
| + | | Quite Weak | ||
|} | |} | ||
<br><br><br><br><br><br><br> | <br><br><br><br><br><br><br> | ||
| - | |||
| - | Single operators or a combination of them, can be also assembled upstream or downstream with respect to constitutive promoter. | + | |
| + | <br><br><br> | ||
| + | Single operators or a combination of them, can be also assembled upstream or downstream with respect to a constitutive promoter. In fact, it is known (Cox et al, 2007) that the position of an operator site plays a crucial role in determining repression effects (i.e. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079041 lambda operators]). In particular, the same operator located upstream of the promoter -35 sequence functions as a weaker repressor than one located downstream of the -10 consensus sequence. | ||
| + | |||
| + | |||
| + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
= Operator site cloning in standard plasmids = | = Operator site cloning in standard plasmids = | ||
| Line 104: | Line 138: | ||
So, we decided to set a new protocol up for the isolation and cloning of single operator sites into standard BioBricks plasmids. | So, we decided to set a new protocol up for the isolation and cloning of single operator sites into standard BioBricks plasmids. | ||
| - | We started from the analysis of an existing | + | We started from the analysis of an existing custom vector into the [http://partsregistry.org/wiki/index.php?title=Part:BBa_J23100 Registry]. In this vector, a RFP gene was cloned, as a reporter, between the SpeI and PstI restriction sites. Thus, this is the only part in the Registry that can be separated from a plasmid with a S/P digestion. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Therefore, once the RFP is isolated with S/P, it could be assembled with an operator sequence digested SpeI- PstI, too. This could allow the isolation of the Operator- RFP sequence from the commercial plasmid and the subsequent religation into a standard plasmid. Then, a final extraction of the RFP gene with a SpeI- PstI digestion would leave the operator site inside the standard plasmid. | |
| - | |||
| - | + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | |
| - | + | ||
| + | = Experimental assesment of LacI operator functionality = | ||
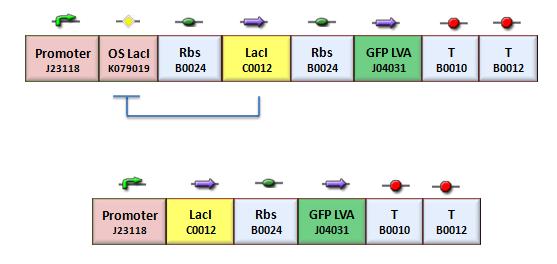
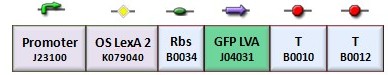
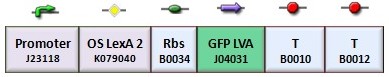
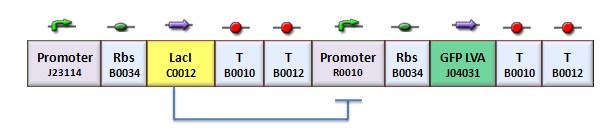
| + | According to the protocol to test operator parts ([https://2008.igem.org/Team:Bologna/Modeling#Procedure_for_Ki-index_identification procedure for K-index identification]), two circuits were assembled (Figure 1): K079020 is the closed-loop configuration where GFP expression is auto-regulated by the synthesis of LacI repressor protein; K079026 is the open-loop configuration lacking the operator site to determinate the maximum fluorescence due to the constitutive promoter. In the latter construct, GFP was spaced from the promoter inserting the LacI gene sequence, to account for the abortive transcriptions. | ||
| - | |||
| - | |||
<br> | <br> | ||
| - | |||
| - | + | [[Image:bba020.jpg|center|thumbnail|400 px|Figure 1. Upper panel: Closed-loop configuration (K079020); lower panel Open-loop configuration (K079026)]] | |
| - | + | <br> | |
| - | + | ||
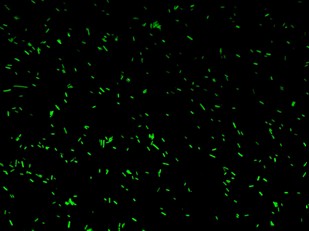
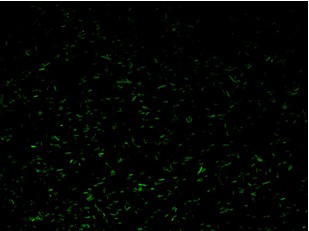
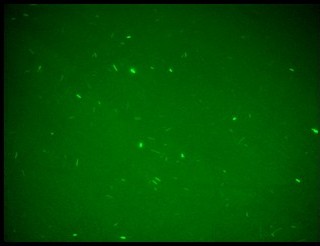
| - | + | K079020 and K079026 were transformed in HL1Blue bacterial cells according to the standard protocol. One colony from each plate was picked up and let grow overnight in LB medium at 37°C. One milliliter for each of the two samples was collected by O/N cultures and spinned at 6000-8000 rpm for three minutes. The supernatant was harvested and the pellet resuspended. Slides were prepared for the fluorescence bacteria image acquisition. For each slide five different views were acquired. Finally, images were elaborated with the Visual Fluo Bacteria Software. Examples of fluorescence bacteria image are shown in Figure 2a-2b. The fluorescence images reveal the repression due to the presence of the Lac operator. | |
| - | The | + | |
| - | + | <br> | |
| - | + | {|align="center" | |
| + | |[[Image:closed.jpg|thumbnail|350 px|Figure 2a. Open-Loop configuration (K079026)]] | ||
| + | |[[Image:open.jpg|thumbnail|350 px|Figure 2b. Closed-Loop configuration (K079020)]] | ||
| + | |} | ||
| - | + | <br> | |
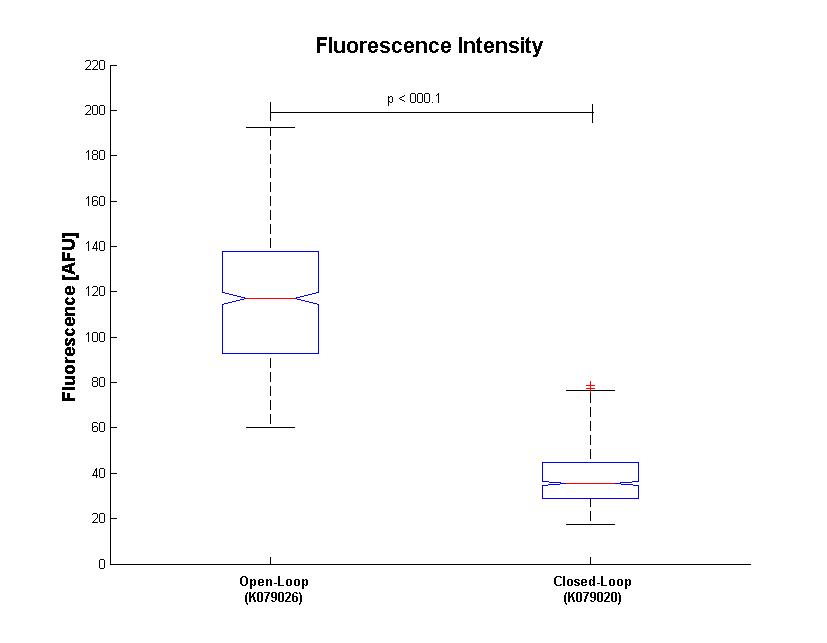
| - | + | Bacterial fluorescence distribution is shown in Figure.3 | |
| - | + | <br> | |
| - | [[Image: | + | [[Image:Box.jpg|thumbnail|center|500 px|Figure 3. Box Plot of bacterium fluorescence (n=683). Max and minimum values are indicated by the horizontal bars. The median and lower and upper quartiles are also indicated.]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <br> | |
| - | + | ||
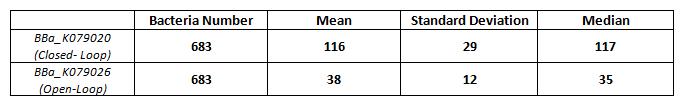
| - | + | Results of fluorescence bacteria analysis by software are reported in tab. | |
| - | + | ||
| - | + | <br> | |
| - | + | [[Image:tab2.jpg|center]] | |
| - | + | <br> | |
| - | + | The open loop and closed loop circuit fluorescence ratio ([https://2008.igem.org/Team:Bologna/Modeling#Procedure_for_Ki-index_identification h factor]) is | |
| - | + | ||
| - | + | <br> | |
| + | |||
| + | [[Image:h2.jpg|center]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | The Ki-index corresponding to this h factor is equal to 4.43 ( [https://2008.igem.org/Team:Bologna/Modeling#Procedure_for_Ki-index_identification see Figure 6 in Modeling 1.5]). The kr-range for the bistability is from 4 to 6 ([https://2008.igem.org/Team:Bologna/Modeling#Bifurcation_analysis see Figure 4 in Modeling 1.4 ]) | ||
| - | |||
| - | |||
[https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
| - | = | + | = Uv-Sensitive Trigger = |
| - | + | ||
| - | + | In the genetic Flip-Flop, the amount of LacI to set ON the memory is induced by an [https://2008.igem.org/Team:Bologna/Modeling#The_genetic_Flip-Flop UV-sensitive trigger ]. Production of LacI molecules by UV induction can be tested by replacing the LacI gene with GFP in the UV trigger, so it is possible to have a relation of the strength of LacI synthesis measuring the value of fluorescence.<br> We realized two new constructs (K079049 and K079050) to test the UV induction. We used two different promoters (J23118 with 1429 strength and J23100 with 2547 strength). These UV test circuits are presented schematically in Figure 4-5. | |
| - | [[Image: | + | |
| - | [[Image: | + | <br> |
| + | |||
| + | [[Image:fig050.jpg|center|thumbnail|388 px| Figure 4. K079050 GFP reporter protein under the control of the J23100 constitutive promoter and LexA 2 operator ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[Image:fig049.jpg|center|thumbnail|388 px| Figure 5. K079049 GFP reporter protein under the control of the J23118 constitutive promoter and LexA 2 operator ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | To test the UV induction we used the [https://2008.igem.org/Team:Bologna/Wetlab#Homemade_UV_Illuminator UV illuminator]. | ||
| + | |||
| + | |||
| + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
| + | |||
| + | = UV Induction = | ||
| + | |||
| + | Plasmids with the K079049 and K079050 constructs have been transformed in DH5alfa bacteria by standard protocol and one colony from the plate was picked up and cultured overnight in 5ml LB medium broth with ampicillin. The day after the culture was diluted in 5ml LB and antibiotic in order to have OD = 0.1 and let it grown for another one hour; after that the culture was divided in five 15ml tubes (1ml of bacteria per tube).<br> | ||
| + | The 1ml volume was used to have a layer of medium enough thin to perform an uniform irradiation.<br> | ||
| + | |||
| + | Tests with difference distances from the lamp (3 and 4 cm) with different exposition times (1, 10 and 15 s) were done. Minimum distance and maximum time were fixed to avoid lethal UV dose and mutagenesis.<br> | ||
| + | |||
| + | After induction by UV the samples were kept for 2 hours in dark by silver paper to increase the RecA and LexA response. The OD was measured and the sample transferred in a 1ml tube, spinned at 6000-8000rpm for three minutes, the supernatant was harvested and the pellet resuspended. Slides were prepared for the fluorescence bacteria image acquisition that were elaborated with the [https://2008.igem.org/Team:Bologna/Software#Visual_Fluo_Bacteria:_a_software_for_the_analysis_of_fluorescence_bacteria_image Visual Fluo Bacteria Software]. <br> | ||
| + | OD was not altered by UV irradiation but we didn't observe GFP. Probably we were not able to find the right time/distance setting. Moreover we are not sure that an uniform irradiation takes place. | ||
| + | |||
| + | |||
| + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
| + | |||
| + | = Hydrogen Peroxide Induction = | ||
| + | |||
| + | Since we have not success with UV induction, we tested the correct functionality of K079049 and K079050 constructs by using hydrogen peroxide induction. It is well known [''Assad et al. 2004''] that hydrogen peroxide is an important reactive oxygen species (ROS). The reaction of hydrogen peroxide with transition metals imposes on cells an oxidative stress that can result in damage to cell components such as proteins, lipids and principally DNA. In E.coli low concentration of H2O2 (1-3mM) results in SOS gene induction [''Imlay and Linn, 1987; Goerlich et alt. 1989'']. | ||
| + | |||
| + | The plasmid contained the LexA Operator was transformed into DH5alfa bacteria according to the standard protocol and one colony was picked up from the plate and let it grown overnight in 15ml tube with 5ml LB medium and ampicillin. Cultured colony was diluted in 5ml LB with antibiotic and 1ul H2O2 (11M) to have medium with 2.2mM concentration of H202 and 0.1 starting OD. | ||
| + | |||
| + | As control, to prove that H2O2 has effect on the K079050, one sample of the same culture was diluted in 5ml LB medium with antibiotic without H2O2. | ||
| + | Additionally, to demonstrate that H202 induction has not interferences with the LacI regulated promoter we also tested the K079029 (Figure 6) that was grown in the LB medium with 2.2mM of H2O2. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[Image:immag2.jpg|center|thumbnail| 500 px| Figure 6. K079029 GFP expression controlled by LacI repressor under the J23114 promoter]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | Tubes were kept to the dark by using silver paper and let them growing for 2 hours at 37°C. 1 ml of bacteria sample has been collected and transferred inside 1ml tube and spinned at 6000-8000 rpm for 3 minutes. The supernatant was discard and the pellet resuspended. Slides were prepared for the fluorescence bacteria image acquisition that were elaborated with the [https://2008.igem.org/Team:Bologna/Software#Visual_Fluo_Bacteria:_a_software_for_the_analysis_of_fluorescence_bacteria_image Visual Fluo Bacteria Software]. | ||
| + | |||
<br> | <br> | ||
| - | |||
{|align="center" | {|align="center" | ||
| - | |[[Image: | + | |[[Image:imm4.jpg|center|thumbnail|300 px|Figure 7. K079050 with H2O2]] |
| - | |[[Image: | + | |[[Image:immag5.jpg|center|thumbnail|300 px|Figure 8. K079050 without H2O2]] |
| + | |[[Image:immag6.jpg|center|thumbnail|300 px|Figure 9. K079029 with H2O2]] | ||
|} | |} | ||
| + | |||
<br> | <br> | ||
| - | + | It is possible to see that GFP is sinthetized only in the construct with Lex A operator. | |
| - | + | In addition to prove that K079050 construct can be used in the Flip-Flop circuit without interference with IPTG, we induced both K079050 and K079029 as positive control for 2 hours with 1mM of IPTG. | |
| + | |||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | {|align="center" | |
| - | + | |[[Image:imm7.jpg|center|thumbnail|300 px| Figure 10. K079029 induced by IPTG]] | |
| + | |[[Image:imm8.jpg|center|thumbnail|300 px| Figure 11. K079050 induced by IPTG]] | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | |||
| + | As expected IPTG was not able to induce the construct with Lex A operator but it induced the circuit with LacI repressed promoter. We conclude that H2O2 induction gives an uniform activity of the regulator promoter (J23100) and the level of promoter activity can be used to set ON the memory. | ||
| + | |||
| + | |||
| + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
= Homemade UV Illuminator = | = Homemade UV Illuminator = | ||
| - | The UV source that we use is a GW6 Sylvania, a lamp that emitt UVC at 253,7nm near the absorption's peak of DNA with an optical output power of 1,6W; to use it [[Team:Bologna/Biosafety#Ethidium_Bromide_and_UV_Rays| | + | The UV source that we use is a GW6 Sylvania, a lamp that emitt UVC at 253,7nm near the absorption's peak of DNA with an optical output power of 1,6W; to use it [[Team:Bologna/Biosafety#Ethidium_Bromide_and_UV_Rays|safely]] we built a box of mdf(medium density-fibreboard) that surrounds the lamp and prevent the UVc leakeage, moreover we use a diaphragm that allows passage only to part of the light, absorbing the remainder. We insert in that box two pierced brackets, that permits to choose the distance between the lamp and the sample and then in accord with the Lambert-Beer law's the exposure time. |
| - | Finally we embedded this structure in a polistyrene box for its handling and greater safety (Figure | + | Finally we embedded this structure in a polistyrene box for its handling and greater safety (Figure 12-13). |
| + | |||
| + | <br> | ||
{|align="center" | {|align="center" | ||
| - | |[[Image:scatola1.jpg|center|thumbnail|300 px|Figure | + | |[[Image:scatola1.jpg|center|thumbnail|300 px|Figure 12]] |
|width=70| | |width=70| | ||
| - | |[[Image:scatola2.jpg|center|thumbnail|300 px| Figure | + | |[[Image:scatola2.jpg|center|thumbnail|300 px| Figure 13]] |
|} | |} | ||
| Line 211: | Line 289: | ||
Our project use UV light for its space selectivity, that gives the possibility to irradiate a target zone | Our project use UV light for its space selectivity, that gives the possibility to irradiate a target zone | ||
without interfering with the other. To do that we build a mold to make a matrix of agarose gel;this | without interfering with the other. To do that we build a mold to make a matrix of agarose gel;this | ||
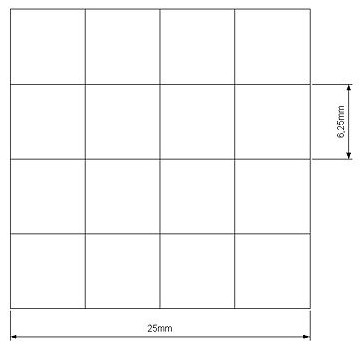
| - | give us a square of 25mm side's with 16 cells inside where we can locate bacteria( Figure | + | give us a square of 25mm side's with 16 cells inside where we can locate bacteria( Figure 14 ). |
| - | [[Image:matrice.jpg|center|thumbnail|300 px| Figure | + | |
| + | <br> | ||
| + | |||
| + | [[Image:matrice.jpg|center|thumbnail|300 px| Figure 14]] | ||
| + | |||
| + | <br> | ||
Once do that is easy with an optical mask, like those used in photolithography for electronics | Once do that is easy with an optical mask, like those used in photolithography for electronics | ||
circuits, stimulate only the selected bacteria.To realize this device we use palsticard because it is | circuits, stimulate only the selected bacteria.To realize this device we use palsticard because it is | ||
| - | very easy to shape and clean( Figure | + | very easy to shape and clean( Figure 15 ). |
The mold is made of two parts that will form a sandwich with the slide: one will create the shape | The mold is made of two parts that will form a sandwich with the slide: one will create the shape | ||
| - | of the gel matrix the other is a cover that that will give the picture into gel .<br> | + | of the gel matrix( Figure 16 ) the other is a cover that that will give the picture into gel. |
| + | |||
| + | <br> | ||
| + | |||
{|align="center" | {|align="center" | ||
| - | |[[Image:imm1.jpg|center|thumbnail|300 px|Figure | + | |[[Image:imm1.jpg|center|thumbnail|300 px|Figure 15]] |
|width=70| | |width=70| | ||
| - | |[[Image:imm2.jpg|center|thumbnail|300 px|Figure | + | |[[Image:imm2.jpg|center|thumbnail|300 px|Figure 16]] |
|} | |} | ||
| + | <br> | ||
| + | |||
| + | The matrix is obtained through the pressure of the cover part over the mold part( Figure 17 ) | ||
| + | |||
| + | <br> | ||
| + | |||
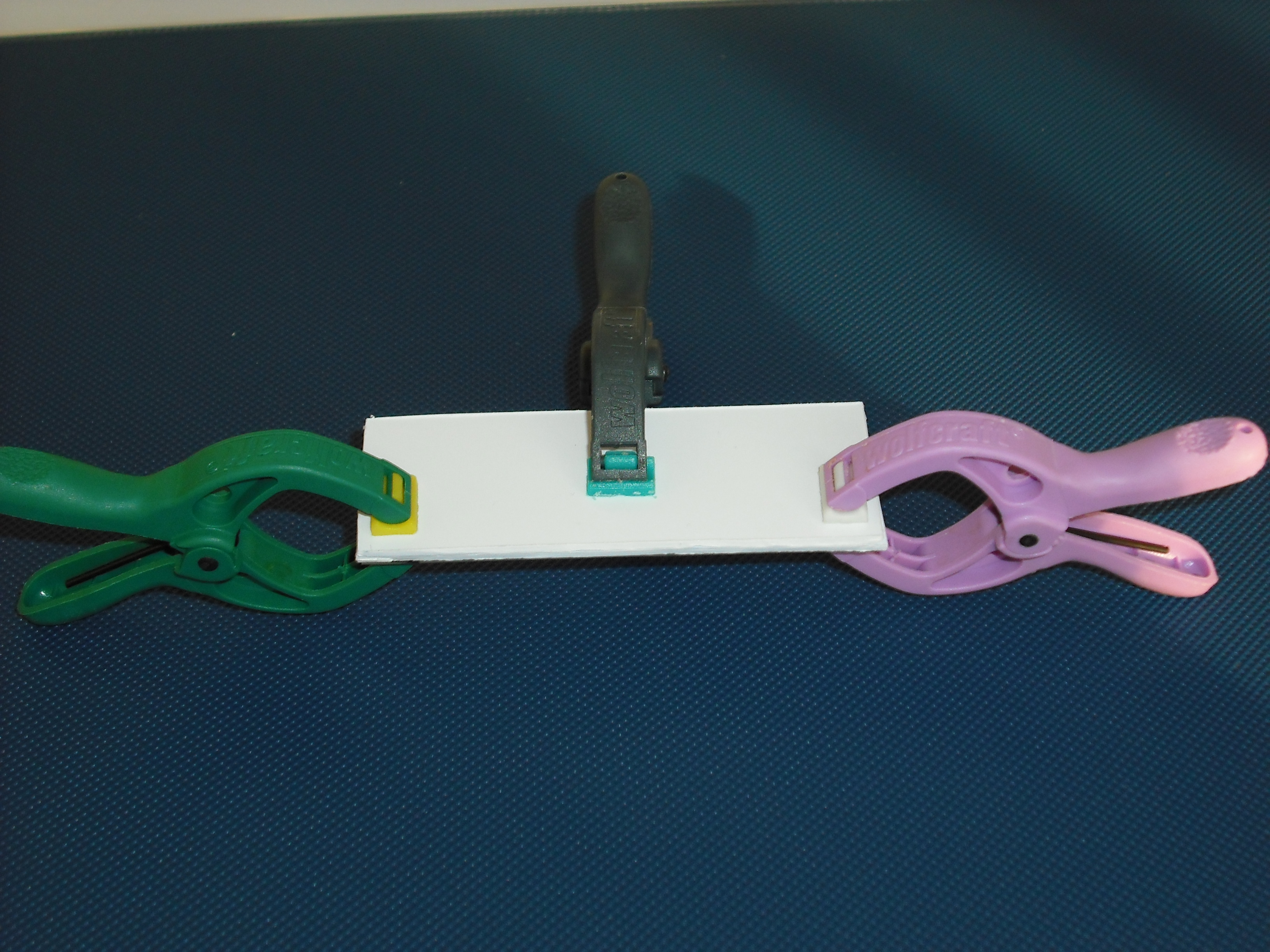
| + | [[Image:pinze.jpg|center|thumbnail|300 px|Figure 17]] | ||
| + | |||
| + | <br> | ||
| - | |||
| - | |||
and that is the result: | and that is the result: | ||
| - | [[Image:imm3.jpg|center|thumbnail|300 px|Figure | + | |
| + | <br> | ||
| + | |||
| + | [[Image:imm3.jpg|center|thumbnail|300 px|Figure 18]] | ||
| + | |||
| + | |||
| + | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
| + | |||
| + | = Bibliography = | ||
| + | |||
| + | *Friedberg EC, Walker GC and Siede W (1995) DNA Repair and Mutagenesis. Academic Press, New York, 698 pp<br> | ||
| + | *Courcelle J, Khoudursky A, Peter B, Brown PO and Hanawalt PC (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64<br> | ||
| + | *Wagner J, Cruz P, Kim SR, Yamada M, Matsui K, Fuchs RP and Nohmi T (1999) The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell 4:281-286<br> | ||
| + | *Mustard JA and Little JW (2000) Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of rec<br> | ||
| + | *Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H and Woodgate R (2000)<br> Identification of additional genes belonging to the lexA regulon in Escherichia coli. Mol Microbiol 35:1560-1572<br> | ||
| + | *Imlay JA and Linn S (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bateriol 169:2967-2976<br> | ||
| + | *Goerlich O, Quillardet P and Hofnung M (1989) Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol 171:6141-6147<br> | ||
| + | *Imlay JA and Linn S (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide<br> | ||
[https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | [https://2008.igem.org/Team:Bologna/Wetlab ''Up''] | ||
Latest revision as of 12:57, 4 December 2008
| HOME | PROJECT | TEAM | SOFTWARE | MODELING | WET LAB | LAB-BOOK | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
Contents |
Introduction
To obtain regulated promoters we synthesized four libraries of operator sequences, respectively for LacI, TetR, cI and LexA repressor proteins. Here is explained how we designed the operator libraries and isolated single operators to clone them in the standard format. Moreover, we built and tested: a LacI regulated promoter to tune promoter activation and a lexA based promoter to study the SOS activation (UV and peroxidase).
At last... Operator sites as BioBricks!
Even though transcriptional regulation still plays a pivotal role in synthetic biology, a modular assembly of regulated promoters and the characterization of their properties has not been formalized, yet. At present state, each promoter in the Registry, though complex, is treated as a “standalone” monolithic element. Thus, the choice of a regulated promoter implies a prefixed sigmoid regulation curve. The only option, using the current BioBricks, is to choose a discretional scaling factor in the transfer of the transcription function to the protein through an appropriate RBS. Critically, the choice of one specific transcription factor limits the choice to just one possible promoter. The assembly of regulated promoters as the combination of modular parts, i. e. unregulated promoters and operators, could permit the rapid design of regulatory elements with fixed characteristics. In fact, promoter transcriptional strength and repressor binding affinity could be independently chosen.
A first step in the rationalization of promoter design was done in the iGEM 2007 with the introduction in the Registry of a family of constitutive promoters. Each family element differs from the others just for few base pairs in -35 and -10 regions, keeping constant the rest of the sequence and giving rise to a different level of transcription strength. We decided to use this valuable work as a platform for a deeper and more general promoter design strategy. To obtain regulated promoters we decided to combine these parts with short regulatory sequences that operate as a binding site for the transcription factor. To this aim, we synthesized four libraries of operator sequences, respectively for [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079045 LacI], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079046 TetR], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079047 cI] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079048 LexA] repressor proteins ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K079045 see details]). In each library, there are three sequences (see Table below) each with a different repressor binding affinity to the repressor protein. Since the libraries were synthetized on pGA18 and pMA Geneart vectors, we isolated each operator with the intention to clone them into BioBrick standard assembly plasmids. The choice of the operator should give a relative fine-tuning of promoter sensitivity to the repressor. We decided to take the Berkley's costitutive promoter library as a good "collection" from which we could select the unregulated promoter, according with the chosen transcriptional strength.
| LacI | ||
| Name | Sequence | Affinity |
| [http://partsregistry.org/Part:BBa_K079017 Lac SymL] | aattgtgagcgctcacaatt | Very Strong |
| [http://partsregistry.org/Part:BBa_K079018 Lac O1] | aattgtgagcggataacaatt | Intermediate |
| [http://partsregistry.org/Part:BBa_K079019 Lac O2] | aaatgtgagcgagtaacaacc | Weak |
| Tet | ||
| Name | Sequence | Affinity |
| [http://partsregistry.org/Part:BBa_K079036 TeT O] | cctatcagtgataga | Strong |
| [http://partsregistry.org/Part:BBa_K079037 TetO-4C] | cctgtcagtgacaga | Intermediate |
| [http://partsregistry.org/Part:BBa_K079038 TetO-wt/4C5G] | cctatcagtgacgga | Weak |
| Lambda | ||
| Name | Sequence | Affinity |
| [http://partsregistry.org/Part:BBa_K079041 Lambda OR1] | tatcaccgccagaggta | Strong/Intermediate cI/Cro |
| [http://partsregistry.org/Part:BBa_K079042 Lambda OR2] | taacaccgtgcgtgttg | Intermediate/Weak cI/Cro |
| [http://partsregistry.org/Part:BBa_K079043 Lambda OR3] | tatcaccgcaagggata | Intermediate/Intermediate cI/Cro |
| Lex | ||
| Name | Sequence | Affinity |
| [http://partsregistry.org/Part:BBa_K079039 LexA 1] | ctgtatatatatacag | Very Strong |
| [http://partsregistry.org/Part:BBa_K079040 LexA 2] | ctgtatgagcatacag | Quite Weak |
Single operators or a combination of them, can be also assembled upstream or downstream with respect to a constitutive promoter. In fact, it is known (Cox et al, 2007) that the position of an operator site plays a crucial role in determining repression effects (i.e. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K079041 lambda operators]). In particular, the same operator located upstream of the promoter -35 sequence functions as a weaker repressor than one located downstream of the -10 consensus sequence.
Operator site cloning in standard plasmids
Operator sites are Dna sequences very small in length (15 to 20 bp) and a restriction digestion for the religation in standard plasmids is not possible with the existing purification kits. In fact, only Dna sequences down to at least 40 bp can be efficiently purified with the standard kits available. Small bands extraction requires laborious condition optimization and hazardous reagents like phenol-chloroform So, we decided to set a new protocol up for the isolation and cloning of single operator sites into standard BioBricks plasmids.
We started from the analysis of an existing custom vector into the [http://partsregistry.org/wiki/index.php?title=Part:BBa_J23100 Registry]. In this vector, a RFP gene was cloned, as a reporter, between the SpeI and PstI restriction sites. Thus, this is the only part in the Registry that can be separated from a plasmid with a S/P digestion.
Therefore, once the RFP is isolated with S/P, it could be assembled with an operator sequence digested SpeI- PstI, too. This could allow the isolation of the Operator- RFP sequence from the commercial plasmid and the subsequent religation into a standard plasmid. Then, a final extraction of the RFP gene with a SpeI- PstI digestion would leave the operator site inside the standard plasmid.
Experimental assesment of LacI operator functionality
According to the protocol to test operator parts (procedure for K-index identification), two circuits were assembled (Figure 1): K079020 is the closed-loop configuration where GFP expression is auto-regulated by the synthesis of LacI repressor protein; K079026 is the open-loop configuration lacking the operator site to determinate the maximum fluorescence due to the constitutive promoter. In the latter construct, GFP was spaced from the promoter inserting the LacI gene sequence, to account for the abortive transcriptions.
K079020 and K079026 were transformed in HL1Blue bacterial cells according to the standard protocol. One colony from each plate was picked up and let grow overnight in LB medium at 37°C. One milliliter for each of the two samples was collected by O/N cultures and spinned at 6000-8000 rpm for three minutes. The supernatant was harvested and the pellet resuspended. Slides were prepared for the fluorescence bacteria image acquisition. For each slide five different views were acquired. Finally, images were elaborated with the Visual Fluo Bacteria Software. Examples of fluorescence bacteria image are shown in Figure 2a-2b. The fluorescence images reveal the repression due to the presence of the Lac operator.
Bacterial fluorescence distribution is shown in Figure.3
Results of fluorescence bacteria analysis by software are reported in tab.
The open loop and closed loop circuit fluorescence ratio (h factor) is
The Ki-index corresponding to this h factor is equal to 4.43 ( see Figure 6 in Modeling 1.5). The kr-range for the bistability is from 4 to 6 (see Figure 4 in Modeling 1.4 )
Uv-Sensitive Trigger
In the genetic Flip-Flop, the amount of LacI to set ON the memory is induced by an UV-sensitive trigger . Production of LacI molecules by UV induction can be tested by replacing the LacI gene with GFP in the UV trigger, so it is possible to have a relation of the strength of LacI synthesis measuring the value of fluorescence.
We realized two new constructs (K079049 and K079050) to test the UV induction. We used two different promoters (J23118 with 1429 strength and J23100 with 2547 strength). These UV test circuits are presented schematically in Figure 4-5.
To test the UV induction we used the UV illuminator.
UV Induction
Plasmids with the K079049 and K079050 constructs have been transformed in DH5alfa bacteria by standard protocol and one colony from the plate was picked up and cultured overnight in 5ml LB medium broth with ampicillin. The day after the culture was diluted in 5ml LB and antibiotic in order to have OD = 0.1 and let it grown for another one hour; after that the culture was divided in five 15ml tubes (1ml of bacteria per tube).
The 1ml volume was used to have a layer of medium enough thin to perform an uniform irradiation.
Tests with difference distances from the lamp (3 and 4 cm) with different exposition times (1, 10 and 15 s) were done. Minimum distance and maximum time were fixed to avoid lethal UV dose and mutagenesis.
After induction by UV the samples were kept for 2 hours in dark by silver paper to increase the RecA and LexA response. The OD was measured and the sample transferred in a 1ml tube, spinned at 6000-8000rpm for three minutes, the supernatant was harvested and the pellet resuspended. Slides were prepared for the fluorescence bacteria image acquisition that were elaborated with the Visual Fluo Bacteria Software.
OD was not altered by UV irradiation but we didn't observe GFP. Probably we were not able to find the right time/distance setting. Moreover we are not sure that an uniform irradiation takes place.
Hydrogen Peroxide Induction
Since we have not success with UV induction, we tested the correct functionality of K079049 and K079050 constructs by using hydrogen peroxide induction. It is well known [Assad et al. 2004] that hydrogen peroxide is an important reactive oxygen species (ROS). The reaction of hydrogen peroxide with transition metals imposes on cells an oxidative stress that can result in damage to cell components such as proteins, lipids and principally DNA. In E.coli low concentration of H2O2 (1-3mM) results in SOS gene induction [Imlay and Linn, 1987; Goerlich et alt. 1989].
The plasmid contained the LexA Operator was transformed into DH5alfa bacteria according to the standard protocol and one colony was picked up from the plate and let it grown overnight in 15ml tube with 5ml LB medium and ampicillin. Cultured colony was diluted in 5ml LB with antibiotic and 1ul H2O2 (11M) to have medium with 2.2mM concentration of H202 and 0.1 starting OD.
As control, to prove that H2O2 has effect on the K079050, one sample of the same culture was diluted in 5ml LB medium with antibiotic without H2O2. Additionally, to demonstrate that H202 induction has not interferences with the LacI regulated promoter we also tested the K079029 (Figure 6) that was grown in the LB medium with 2.2mM of H2O2.
Tubes were kept to the dark by using silver paper and let them growing for 2 hours at 37°C. 1 ml of bacteria sample has been collected and transferred inside 1ml tube and spinned at 6000-8000 rpm for 3 minutes. The supernatant was discard and the pellet resuspended. Slides were prepared for the fluorescence bacteria image acquisition that were elaborated with the Visual Fluo Bacteria Software.
It is possible to see that GFP is sinthetized only in the construct with Lex A operator.
In addition to prove that K079050 construct can be used in the Flip-Flop circuit without interference with IPTG, we induced both K079050 and K079029 as positive control for 2 hours with 1mM of IPTG.
As expected IPTG was not able to induce the construct with Lex A operator but it induced the circuit with LacI repressed promoter. We conclude that H2O2 induction gives an uniform activity of the regulator promoter (J23100) and the level of promoter activity can be used to set ON the memory.
Homemade UV Illuminator
The UV source that we use is a GW6 Sylvania, a lamp that emitt UVC at 253,7nm near the absorption's peak of DNA with an optical output power of 1,6W; to use it safely we built a box of mdf(medium density-fibreboard) that surrounds the lamp and prevent the UVc leakeage, moreover we use a diaphragm that allows passage only to part of the light, absorbing the remainder. We insert in that box two pierced brackets, that permits to choose the distance between the lamp and the sample and then in accord with the Lambert-Beer law's the exposure time. Finally we embedded this structure in a polistyrene box for its handling and greater safety (Figure 12-13).
Gel matrix
Our project use UV light for its space selectivity, that gives the possibility to irradiate a target zone without interfering with the other. To do that we build a mold to make a matrix of agarose gel;this give us a square of 25mm side's with 16 cells inside where we can locate bacteria( Figure 14 ).
Once do that is easy with an optical mask, like those used in photolithography for electronics circuits, stimulate only the selected bacteria.To realize this device we use palsticard because it is very easy to shape and clean( Figure 15 ). The mold is made of two parts that will form a sandwich with the slide: one will create the shape of the gel matrix( Figure 16 ) the other is a cover that that will give the picture into gel.
The matrix is obtained through the pressure of the cover part over the mold part( Figure 17 )
and that is the result:
Bibliography
- Friedberg EC, Walker GC and Siede W (1995) DNA Repair and Mutagenesis. Academic Press, New York, 698 pp
- Courcelle J, Khoudursky A, Peter B, Brown PO and Hanawalt PC (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64
- Wagner J, Cruz P, Kim SR, Yamada M, Matsui K, Fuchs RP and Nohmi T (1999) The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell 4:281-286
- Mustard JA and Little JW (2000) Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of rec
- Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H and Woodgate R (2000)
Identification of additional genes belonging to the lexA regulon in Escherichia coli. Mol Microbiol 35:1560-1572
- Imlay JA and Linn S (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bateriol 169:2967-2976
- Goerlich O, Quillardet P and Hofnung M (1989) Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol 171:6141-6147
- Imlay JA and Linn S (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide
 "
"