Team:TUDelft/Temperature testing
From 2008.igem.org
Bavandenberg (Talk | contribs) (→Thermosensitive constructs) |
Bavandenberg (Talk | contribs) (→Controls) |
||
| Line 85: | Line 85: | ||
[[Image:TUDelft_compositecontrol.jpg | 200px|center]] | [[Image:TUDelft_compositecontrol.jpg | 200px|center]] | ||
| + | <div class="center"> | ||
{| border="1" | {| border="1" | ||
|+ <b>Table 2: Composite parts with thermosensitive region</b> | |+ <b>Table 2: Composite parts with thermosensitive region</b> | ||
| Line 96: | Line 97: | ||
|} | |} | ||
| + | </div> | ||
==Practical Analysis== | ==Practical Analysis== | ||
Revision as of 14:13, 29 October 2008
Contents |
Testing
Introduction
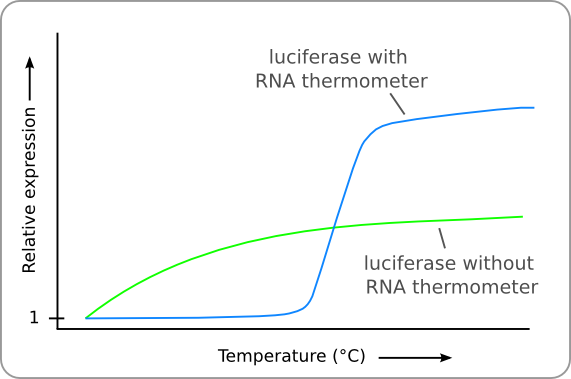
To test the functionality of our RNA-thermometer, we will need a reporter to see whether different temperatures give different translation. Assuming the system is not an on/off switch system, we will need a reporter which we can quantify. After some discussion with our advisors and some people from the Biotechnology Department, we decided luciferase would be the best system to use as a reporter, as it can be quantified. As a negative control for our RNA-thermometer, we would need a luciferase without a RNA-thermometer in front of it. Using the activity of this construct, we would then be able to calculate the normalized activity of the construct including our RNA-thermometer, as displayed in graph 1.
Graph 1: Effect of temperature on normalized expression of Luciferase behind a thermosensitive promoter.
Thermosensitive constructs
We will use the following construct, being the parts as mentioned in table 1. We have designed various thermo sensitive sequences to obtain thermometer RNA allowing translation at different temperatures and to see if we can find trends in effects of these altered sequences on the overall working of the RNA. These specifications can also be found in table 1.
| Expected T (°C) | Source | Promoter | RNA-thermo + RBS | Reporter | 2xTerminator |
|---|---|---|---|---|---|
| 42 | ROSE | R0080 | K115001 | J52008 | B0015 |
| 37 | FourU | R0080 | K115002 | J52008 | B0015 |
| 37 | PrfA | R0080 | K115003 | J52008 | B0015 |
| 42 | ROSE | R0080 | K115008 | J52008 | B0015 |
| 37 | FourU | R0080 | K115009 | J52008 | B0015 |
| 27 | ROSE | R0080 | K115016 | J52008 | B0015 |
| 32 | ROSE | R0080 | K115017 | J52008 | B0015 |
| 37 | ROSE | R0080 | K115020 | J52008 | B0015 |
Controls
As we want to calculate normalized activities, we will also require a negative control, without the thermometer region. This control will require a normal RBS instead. Although the RBS of the thermo sensitive RNAs is not equal to this, it is possible to correct for this mathematically. This is visible by the constant level of the thermosensitive construct above 42oC in graph 1. The construct can be found in table 2.
| Promoter | RBS | Reporter | 2xTerminator |
|---|---|---|---|
| R0080 | B0034 | J52008 | B0015 |
Practical Analysis
The protocol for the validation will be as follows:
- Grow culture without induction.
- Split cultures into different temperatures and induce promoter.
- Grow for 3 hours.
- Perform luciferase assay by use of a kit.
It is important to note that because the growing speed of colonies is greatly influenced by temperature, and thus also the production of mRNA/protein, the activity at each temperature has to be normalized by a negative control grown at the same temperature.
We've displayed schematically what we would expect to see in luciferase activity in table 3.
| Construct | Induced | 20oC | 37oC |
|---|---|---|---|
| Normal RBS | Yes | + | + |
| Thermosensitive RBS | Yes | - | + |
| Thermosensitive RBS | No | - | - |
 "
"