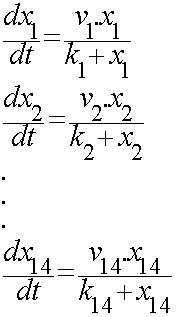Team:TUDelft/Color modeling
From 2008.igem.org
| Line 9: | Line 9: | ||
For the 14 reactions of enzyme-subtrate the Michaelis-Menten kinetics is applied and we have the mass action kinetics for the three degradations. | For the 14 reactions of enzyme-subtrate the Michaelis-Menten kinetics is applied and we have the mass action kinetics for the three degradations. | ||
According to the kinetic laws there are 14 differential equations for enzyme-subtrate reactions and three for degradations which could be constructed as: | According to the kinetic laws there are 14 differential equations for enzyme-subtrate reactions and three for degradations which could be constructed as: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | [[Image:Eq1.gif |120px]] | ||
Revision as of 14:39, 21 August 2008
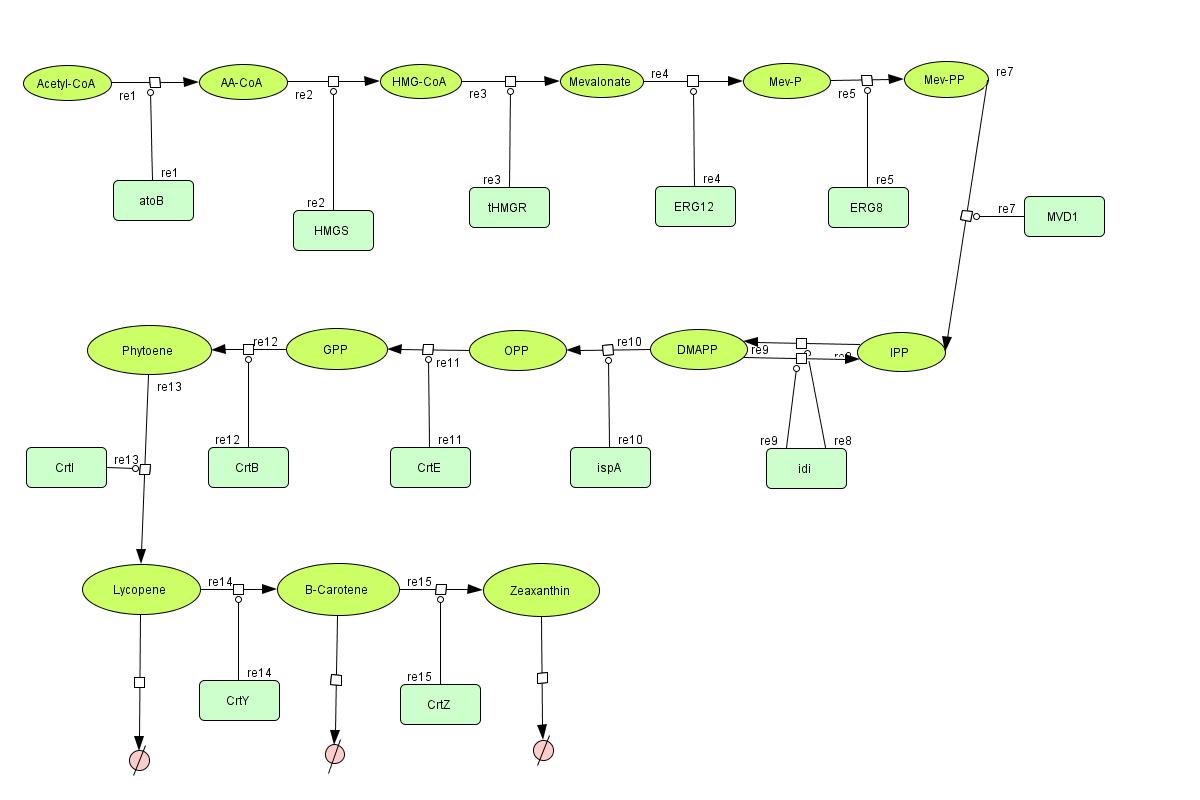
In the output we use mevalonate and GPP pathways together to produce red, orange and yellow colors. The biosynthetic model is bulit in CellDesigner™. There are 14 subtrates and 13 enzymes in total. The first subtrate is Acetyl-CoA which is provided to the pathway in a constant level. The last three subtrates are the color products and are consuming so we have degradation links for them. Lycopene, B-carotene and Zeaxanthin give red, orange and yellow colors respectively. For the 14 reactions of enzyme-subtrate the Michaelis-Menten kinetics is applied and we have the mass action kinetics for the three degradations. According to the kinetic laws there are 14 differential equations for enzyme-subtrate reactions and three for degradations which could be constructed as:
 "
"