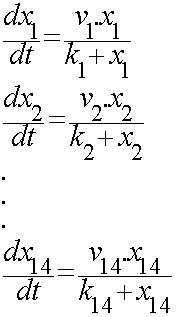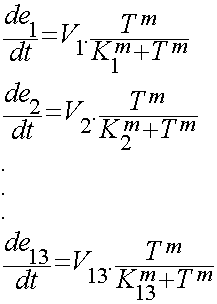Team:TUDelft/Color modeling
From 2008.igem.org
>> work in progress
Contents |
The Model
The model of biothermometer is included an input and output. The input of the model is the enzyme synthesis controlled by the temperature and the output is color production. The challenges for the modeling part are:
- Finding a suitable model for a temperature dependence synthesis for the color producing enzymes.
- Solving the Michaelis-Menten kinetic equations to obtain the dynamics of pathway intermediates.
The procedure for modeling was started with the output part, color pathway, and continued to the enzyme synthesis which can be controlled by temperature.
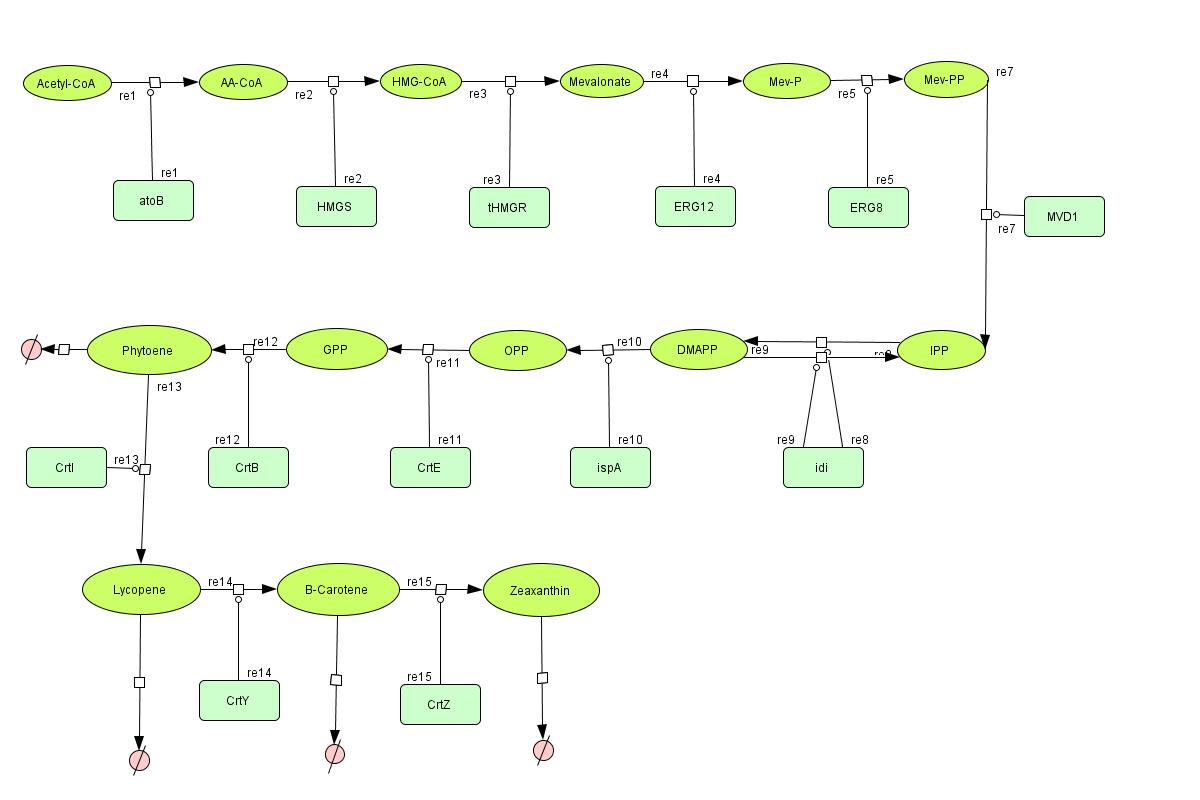
Color Modeling
In the output we use mevalonate and GPP pathways together to produce red, orange and yellow colors. The biosynthetic model is built in CellDesigner™. There are 14 substrates and 13 enzymes in total. The first substrate is Acetyl-CoA which is provided to the pathway in a constant level. The last three substrates are the color products and are consuming so we have degradation links for them. Lycopene, B-carotene and Zeaxanthin give red, orange and yellow colors respectively.
Kinetic Equations
For the 14 reactions of enzyme-substrate the Michaelis-Menten kinetics is applied and we have the mass action kinetics for the three degradations. According to the kinetic laws there are 14 differential equations for enzyme-substrate reactions and three for degradations which could be constructed as:
x1 to x14 are the subtrates.
We used hill type model for enzyme temperature relations where e1 to e13 are the enzymes. Afterwards the in CellDesigner™ file was imported into Matlab. For this procedure many softwares were tested but finally the synthetic biology workbench site used to convert the SBML model into m file. This m file was the base part for the modeling in Matlab. In this m file all the related differential equations were defined as a function with the time span and initial condition as the input. The reactions coefficients are defined by some not systemically acquired numbers to give an estimation for the reactions. These differential equations were solved by Matlab ODE function. Due to the weaknesses in CellDesigner for defining the SBML file the converted file had many problems. To solve these problems the m file was modified.
Initial solution for ODEs
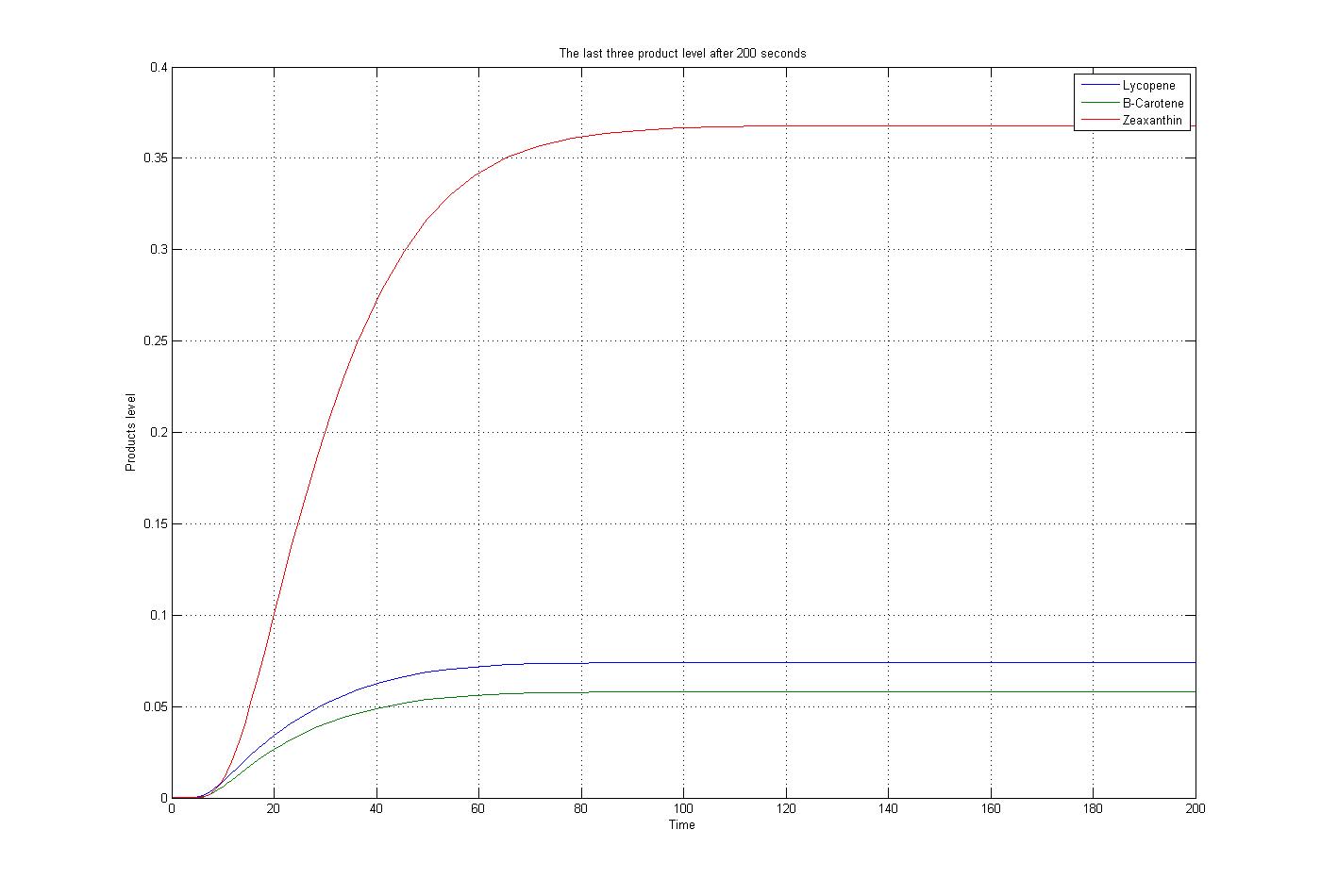
The m file function, as mentioned before, was solved by the ode15s function of Matlab. To do this procedure some assumptions were applied. The first substrate, Acetyl-CoA, assumed to produced and consumed with the same ratio. Also for simplification the level of enzymes assumed to be constant. The reaction coefficients are defined the same for all reaction with some non-accurate numbers to give an overview about the reactions. The results for these assumptions and specific boundary condition and time span of 200 seconds were plotted. These results show the steady state situation for the last three products after 200 seconds. The plot is:
Effects of the temperature on enzymes
Lab experiments for estimating parameters
Parameters calculations
Bifurcation analysis
To have a stable system it is important to do a bifurcation analysis to find the stable ranges for different reaction parameters. This procedure has been done by defining the parameters and substrates concentration as symbolic values in Matlab. Afterwards by these symbolic values the reactions are defined in symbolic format. By these symbolic reactions a vector for reaction ratios for different substrates can be derived. Afterwards by applying a specific steady state, which has been derived from previous step but by applying the initial parameters that have been found from the databases, and changing the reactions parameters in the range that has been found from the data base different reaction vectors were obtained. The changes in the parameter ranges has been define by a random selection in a normal distribution between 0.001 to 0.0374. Subsequently, different vectors for reactions can be derived from this step. The Jacobian applied on each vector where the derivations were respect to the substrate and product concentrations. To conclude the stability for each situation it is necessary that all the eigenvalues of each Jacobian matrix be smaller than zero. If the condition is satisfied then the reactions and pathways are in the stable situation. By this method it is possible to find an approximate ranges for the reaction parameters.
The next step is to find the coefficients of V and K for the both set of equations. In the subtrate equations we have Vmax=Kcat . E0 where Kcat is the turnover number and E0 is the enzyme's initial concentration.
Experiment Vs Theory
... to be continued
 "
"