Team:TUDelft/Temperature design2
From 2008.igem.org
Bavandenberg (Talk | contribs) (→Design approach) |
Bavandenberg (Talk | contribs) (→Design approach) |
||
| Line 27: | Line 27: | ||
Image:Tudelft_design_starting_point_hairpin.png | Temperature sensitive hairpin | Image:Tudelft_design_starting_point_hairpin.png | Temperature sensitive hairpin | ||
</gallery> | </gallery> | ||
| - | |||
| - | |||
[[Image:Tudelft_summed_unaligned_noloop.png | thumb]] | [[Image:Tudelft_summed_unaligned_noloop.png | thumb]] | ||
| + | |||
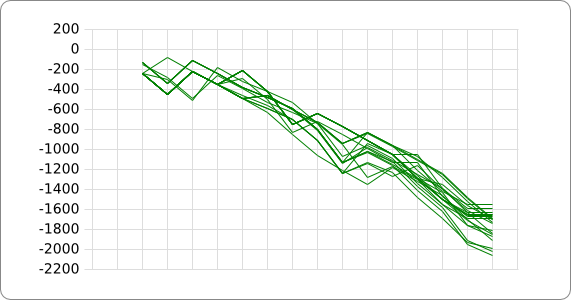
| + | The analysis of the temperature sensitive part of the ROSE RNA thermometer has shown a trend in the binding forces of the base pairs that is common to the different thermometers as also shown in the figures below. | ||
The design of an RNA thermometer is now based on the following requirements: | The design of an RNA thermometer is now based on the following requirements: | ||
Revision as of 13:40, 2 September 2008
>> Work in progress
Contents |
Parts Design II
BBa:K115016, BBa:K115017, BBa:K115018, BBa:K115019
Changing the temperature threshold
The principle of the RNA thermometer is based on base pairing between the nucleotides in the Shine Dalgarno region. At a certain temperature the RNA is folded into a structure in which base pairing nucleotides make the Shine Dalgarno region unreachable for the ribosome. With a rise in temperature, the binding forces between the base pairing nucleotides decrease and above a certain threshold temperature the binding forces between the base pairs are to weak to hold the base pairing nucleotides together, causing the RNA to (partially) unfold. The binding between the base pairing nucleotides let loose, exposing the Shine-Dalgarno region and thereby enabling the ribosome to initiate translation.
When only looking at this principle, an RNA thermometer with a different temperature threshold can be designed by simply increasing or decreasing the binding forces of the base-pairing nucleotides in the Shine-Dalgarno region, shifting the temperature threshold to a higher or lower temperature respectively (fig). Increasing the binding forces could be achieved by incorporating base pairing C and G nucleotides, which bind relatively strong forming a stable helix. Decreasing the binding forces could be done by introducing less strong binding nucleotide base-pairs, such as A-U and G-U base pairs, forming a less stable helix.
But things are not that easy. At first, adding mutations in order to enforce or weaken the temperature sensitive region can also cause the RNA to fold into a completely different structure. This way it can lose its function as an RNA thermometer. Secondly, there are some constraints to the possible mutations; the start codon and the Shine Dalgarno sequence must off course remain unaltered and in case of a standard biobrick the scar must also be part of the temperature sensitive region (see the scar problem). Third, although it seems that only a thermosensitive hairpin is needed to have a functioning RNA thermometer (as described in the analysis section) there could very well be more factors that are of influence on the functioning and temperature threshold of the RNA thermometer that are still unknown.
Design approach
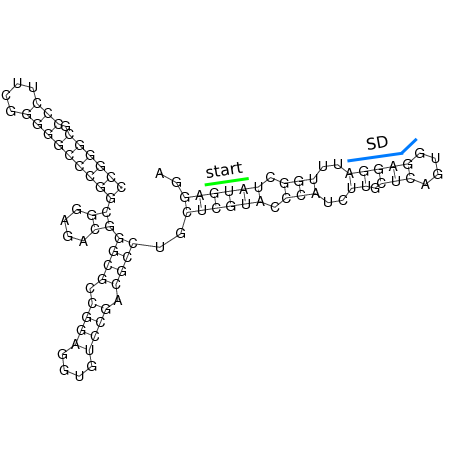
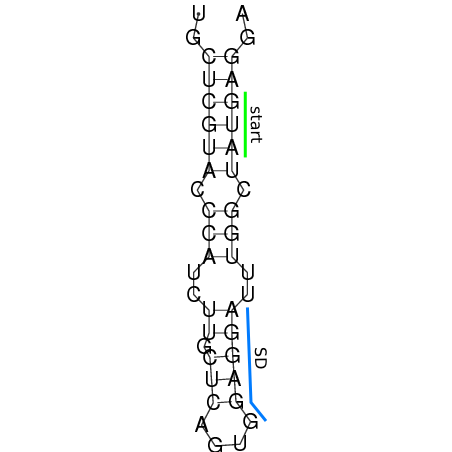
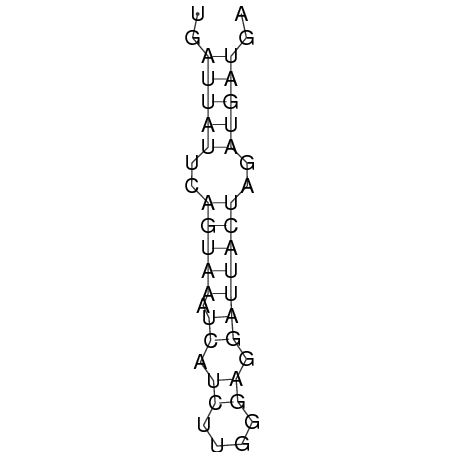
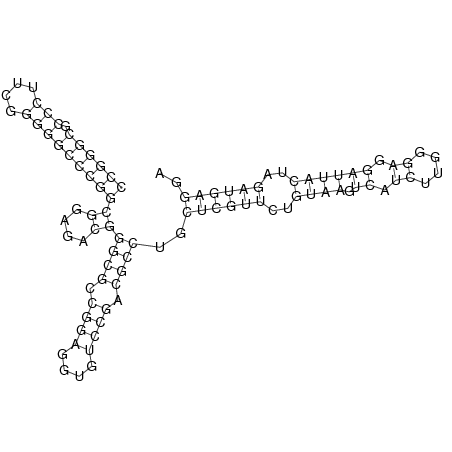
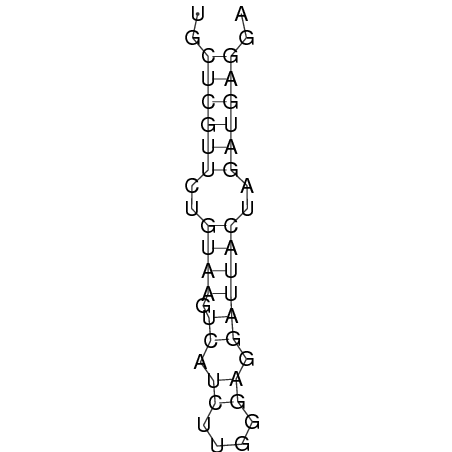
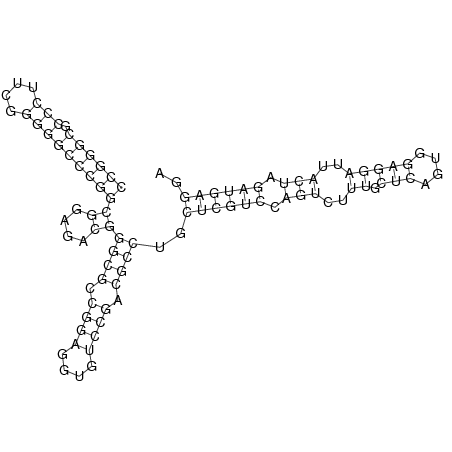
In order to design an RNA thermometer with a different temperature threshold an existing RNA thermometer is taken as a starting point. The one chosen is a ROSE RNA thermometer from Bradyrhizobium japonicum USDA 110 (NC_004463) residing at location 5784144-5784239 within the genome which is at the 5' side of the gene hspB. The reason that a sequence of the ROSE family is chosen instead of one of the FourU family is that there are more ROSE than FourU sequences available which can be used for the analysis of the temperature sensitive part of the RNA thermometer. The reason that a sequence of the ROSE family is chosen instead of one of the PrfA family is that there are more papers that provide information about the working and effects of mutation on ROSE RNA thermometers than on PrfA RNA thermometers. There is no specific reason for the choice of this ROSE RNA thermometer, this is just a ROSE RNA thermometers taken from the Rfam database. The figure below shows the secondary structure of the whole RNA thermometer and the temperature sensitive hairpin as predicted by RNAfold.
The analysis of the temperature sensitive part of the ROSE RNA thermometer has shown a trend in the binding forces of the base pairs that is common to the different thermometers as also shown in the figures below.
The design of an RNA thermometer is now based on the following requirements:
- The sequence before the temperature sensitive hairpin remains unaltered, so mutations are only allowed within the temperature sensitive hairpin.
- The number of nucleotides in the temperature sensitive hairpin is fixed.
- The overall structure of the temperature sensitive hairpin must remain the same. This means that the loop at the end should be of the same size and at the same position. There are no restrictions to the number and locations of internal loops and bulges within the temperature sensitive hairpin.
- The start codon, SD sequence, and scar sequence are fixed.
(TODO: explain the different points)
A design can now be made by fitting the stacking energies plot of the designed hairpin to the found stacking energies trend as given in the figure above, taking into account the just mentioned requirements.
The design of a temperature sensitive hairpin that 'switches' at a certain temperature can be made using the following heuristic:
- Choose a desired switching temperature.
- Take a sequence with the predefined nucleotides (start codon, SD seqeunce, and the scar) present at the correct locations, the rest of the nucleotides can be filled at random.
- Predict the secondary structure of the temperature to check if it corresponds to the required structure. If not, go to step 2.
- Compare the stacking energies plot to the stacking energies trend and calculate the distance between the two plots. If it is the smallest distance thus far, remember the sequence as the best one thus far and go back to step 2 to give it another try. Otherwise go back to step 2.
Design of a 27, 32, and 37 switch
Only one hairpin designs
References
- ^ De Smit M H, Van Duijn J (1990). "Secondary structure of the ribosome binding site determines translation efficiency: A quantitative analysis". PNAS, 1990-10, vol.87, no.19, 7668-7672. PMID:2217199
- ^ Hoe N P, Goguen J D (1993). "Temperature sensing in Yersinia pestis: Translation of the LcrF activator protein is thermally regulated". J Bacteriol, 1993 December, 175(24), 7901-7909. PMID:7504666
- ^ Chowdhurry S, Maris C, Allain F H T, Narberhaus F (2006). "Molecular basis for temperature sensing by an RNA thermometer". The EMBO Journal, 2006, 25, 2487–2497. PMID:16710302
- ^ Nocker A, Hausherr T, Balsiger S, Krstulovic N, Hennecke H, Narberhaus F (2001). "A mRNA-based thermosensor controls expression of rhizobial heat shock genes". Nucleic Acids Research, 2001 December 1, 29(23):4800-4807. PMID:11726689
- ^ Balsiger S, Ragaz C, Baron C, Narberhaus F. "Replicon-specific regulation of small heat shock genes in Agrobacterium tumefaciens". Journal of Bacteriology, October 2004, p. 6824-6829, Vol.186, No.20. PMID:15466035
- ^ Waldminghaus T, Heidrich N, Branti S, Narberhaus F (2007). "FourU: a novel type of RNA thermometer in Salmonella". Molecular Microbiology, Volume 65, Issue 2, 413-424. PMID:17630972
- ^ Johansson J, Mandin P, Renzoni A, Chiaruttinni C, Springer M, Cossart P. "An RNA thermosensor controls expression of virulance genes in Listeria monocytogenes". Cell , Volume 110 , Issue 5 , 551-561. PMID:12230973
 "
"