Team:TUDelft/Color testing
From 2008.igem.org
Contents |
Color pathway testing
Correct genes testing
The first indication of whether obtaining of the genes was a succes, is the correct size of DNA product on a gel. DNA sequencing will provide final certainty of the correct genes.
Correct activity testing
The ultimate proof of activity of the pathway would obviously be a colored Escherichia coli colony. However, to identify individual working enzymes, we'd need specific enzymatic assays. Furthermore this is useful, as some enzymatic assays could yield parameters for the modeling guys. Due to a lack of time, we've not looked any further into these assays.
Results
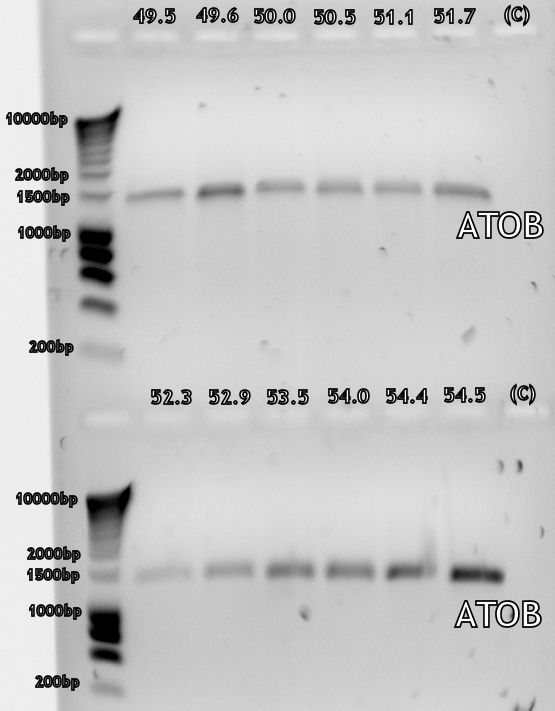
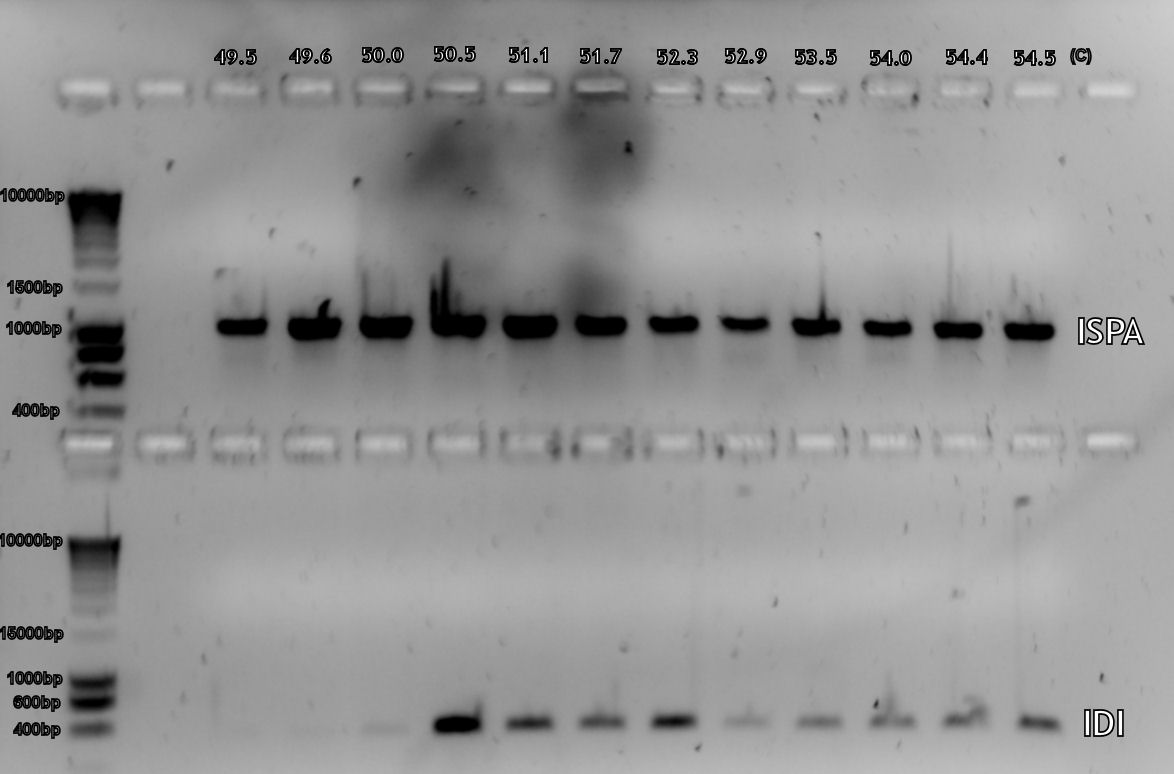
After we had our first succesful PCR on the genome of E. coli with Taq polymerase (figure 1) we tried to repeat it using Pfx polymerase, but this didn't work (figure 2). In the end we decided to repeat the experiment with Taq polymerase as the desired sequences were not very long, thus the chance for mutations due to the lack of proofreading of Taq polymerase was small. In the end the sequences will be verified through DNA sequencing.
The first gradient PCR have been performed on the genes, the E. coli genes all worked correctly (see lab notebook entry of August 19th), while still some work has to be done on the S. cerevisiae primers (August 20th). The next step will be to PCR the E. coli genes again, with the ideal annealing temperature, using Pfx polymerase instead of Taq. The Pfx enzyme has proofreading and so is less likely to make a mistake within the genes. The gradient PCR on S. cerevisiae cDNA will be repeated and optimized.
 "
"