Imperial College/23 September 2008
From 2008.igem.org
(Difference between revisions)
(New page: ==Dry Lab== *Modelling Motility **Done up the code to model the cell's trajectory using least square curve fit method. **Fitted cell trajectories of 100908 Video 15. Results look promisin...) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Imperial/StartPage}}__NOTOC__ | ||
| + | {| cellpadding="10" border="0" | ||
| + | |- valign="top" | ||
| + | |{{#calendar: title=Imperial_College |year=2008 | month=08}} | ||
| + | |{{#calendar: title=Imperial_College |year=2008 | month=09}} | ||
| + | |{{#calendar: title=Imperial_College |year=2008 | month=10}} | ||
| + | | rowspan="2" bgcolor=#ffffff width="100%" | | ||
| + | |} | ||
| + | |||
| + | =23 September 2008= | ||
| + | ==Wet Lab== | ||
| + | ===Single Colony PCR=== | ||
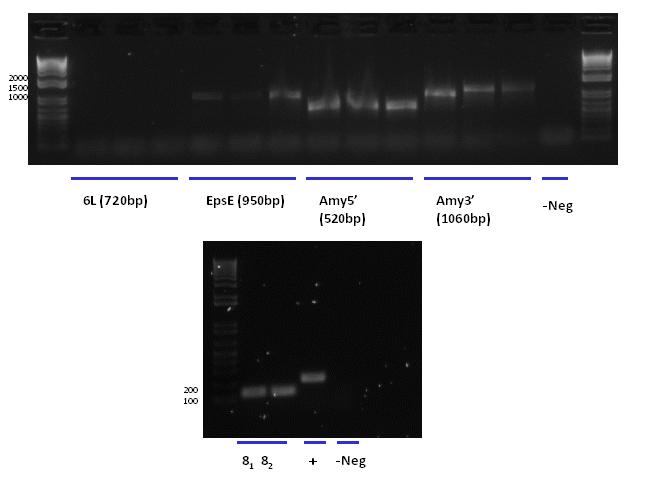
| + | [[Image:PCR-23.PNG|center|500px]] | ||
| + | *In order to verify the ligations of parts (from geneart and PCR products) into the biobrick vector AK3 we have carried out a series of single colony PCRs using the primers Psb. | ||
| + | *The conditions used were as follows: | ||
| + | **1 cycle - 95<sup>o</sup>C for 30 seconds | ||
| + | **30 cycles - 95<sup>o</sup>C for 30 seconds, 60<sup>o</sup>C for 30 seconds,72<sup>o</sup>C for 30 seconds | ||
| + | **1 cycle - 72<sup>o</sup>C for 2 minutes | ||
| + | *The numbers of the ligations correspond to the following ligation reactions: | ||
| + | **6 L (from geneart construct 6) = LipA-Elastin | ||
| + | **EpsE (from geneart construct 3) = EpsE gene | ||
| + | **AmyE5' (from PCR products) | ||
| + | **AmyE3' (from PCR products) | ||
| + | **8<sub>1</sub> (from geneart constructs 6, PCR using mini DNA) = pGsiB-gsiB | ||
| + | **8<sub>2</sub> (from geneart constructs 6, PCR using mini DNA) = pGsiB-gsiB | ||
| + | **Negative contains no DNA, | ||
| + | *The results show positive results for EpsE, Amy5',AmyE3', 8 (pGsiB-gsiB) | ||
| + | *The result from AmyE3' is less clear, it appears that the 1st of the three is the correct size but all will be checked by mini-preping and then digestion. | ||
| + | *There was no contamination in the negative control. | ||
| + | |||
| + | ===Miniprep Digestion=== | ||
| + | |||
| + | *All minpreps from yesterday digested with ''Eco''RI and ''Pst''I | ||
| + | |||
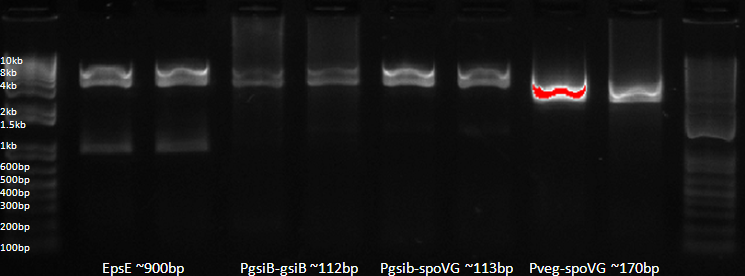
| + | [[Image:Digest23-9.png|center|500px]] | ||
| + | |||
| + | *Each lane is a separate miniprep (2 minipreps of each transformation). | ||
| + | *Band from the EpsE digests is approximately correctly, although this is also approxiamtely the size of GFP-Termiantor and RFP-Terminator. In particular, the pSB1AK3 vector containing RFP-T was used to provide vector for the ligation. | ||
| + | *All other mini-preps do not appear to have inserts! The gel had been imaged earlier and had also shown no evidence of inserts. | ||
| + | *It should also be noted that the vector in all the minipreps appears to be the wrong size! | ||
| + | |||
==Dry Lab== | ==Dry Lab== | ||
*Modelling Motility | *Modelling Motility | ||
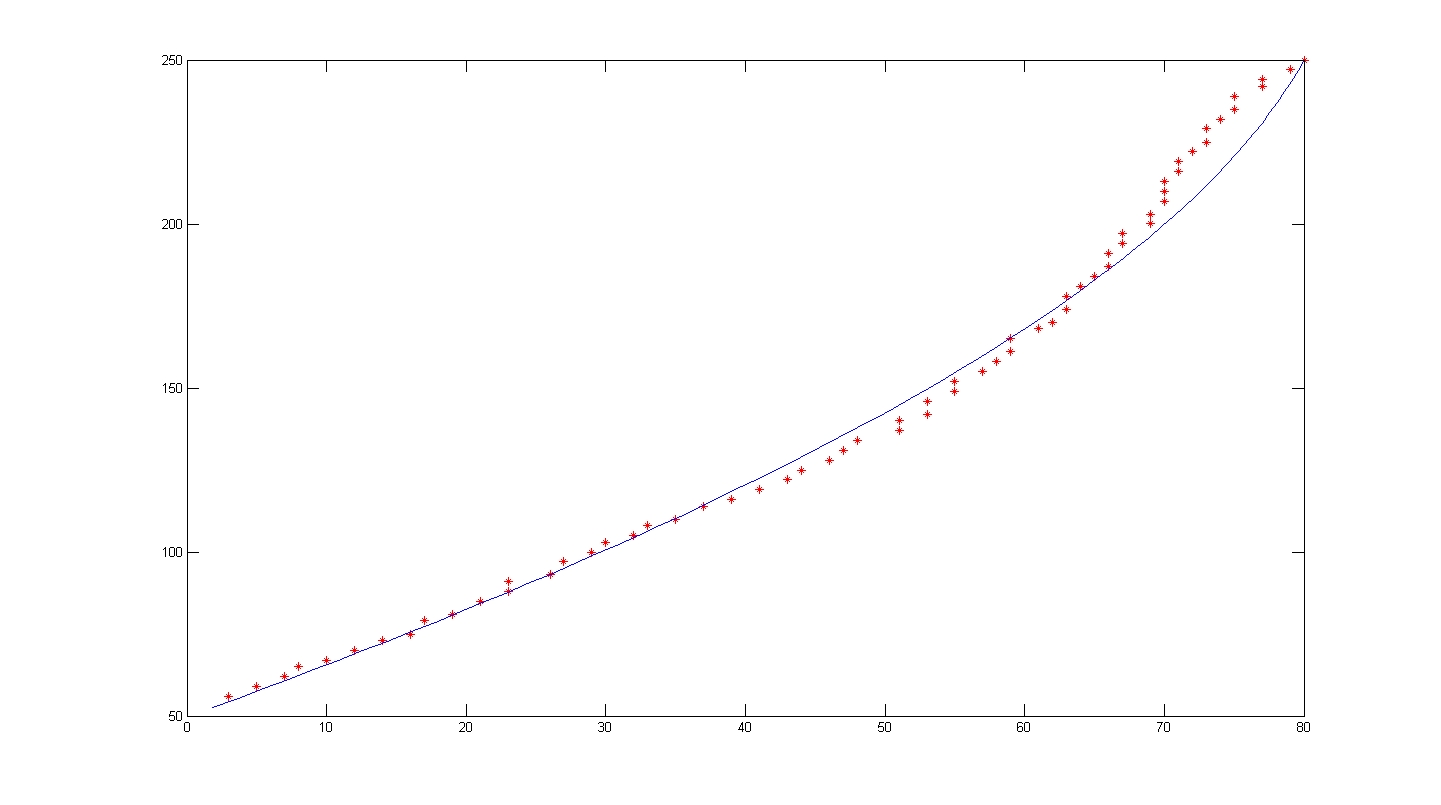
**Done up the code to model the cell's trajectory using least square curve fit method. | **Done up the code to model the cell's trajectory using least square curve fit method. | ||
**Fitted cell trajectories of 100908 Video 15. Results look promising: | **Fitted cell trajectories of 100908 Video 15. Results look promising: | ||
| + | [[Image:V15Cell1.JPG|thumb|500px|center|Video 15 Cell 1]] | ||
| + | [[Image:V15Cell5.jpg|thumb|500px|center|Video 15 Cell 5]] | ||
| + | <br> | ||
| + | {{Imperial/EndPage|Notebook|Notebook}} | ||
Latest revision as of 20:48, 28 October 2008
23 September 2008Wet LabSingle Colony PCR
Miniprep Digestion
Dry Lab
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"