Imperial College/4 September 2008
From 2008.igem.org
(Difference between revisions)
m (→Wetlab) |
|||
| Line 10: | Line 10: | ||
=4 September 2008= | =4 September 2008= | ||
==Wetlab== | ==Wetlab== | ||
| + | |||
| + | ===Cloning=== | ||
| + | |||
| + | *Pfu PCR reactions for LacI gene, Aad9 gene and AmyE 3' Integration Sequence set up using the standard conditions and reagents [http://openwetware.org/wiki/IGEM:IMPERIAL/2008/Prototype/Wetlab/PCR]; LacI and AmyE 3' at 52°C for the first 10 cycles and 65°C for the last 20, Aad9 at 52°C then 60°C. 5μl of each PCR reaction run on a gel - results below | ||
| + | *CAT biobrick digested with ''EcoRI'' and ''SpeI'' ready for assembly with the terminator | ||
| + | *Termiantor biobrick cut with ''EcoRI'' and ''XbaI'' ready for assembly with CAT | ||
| + | *GFP-Termiantor biobrick cut again with ''XbaI'' and ''SpeI'' to produce biobrick vectors for our PCR clones | ||
| + | *GFP-Termiantor biobrick cut again with ''EcoRI'' and ''SpeI'' to produce biobrick vectors for our GeneArt produced clones | ||
| + | |||
===Preparation of Electrocompetent ''E. coli'' Cells=== | ===Preparation of Electrocompetent ''E. coli'' Cells=== | ||
We have prepared 42 aliquots of electrocompetent XL1 Blue ''E. coli'' cells using [http://openwetware.org/wiki/IGEM:IMPERIAL/2008/New/Protocols/XL1-Blue_preparation the relevant protocol] listed on our Protocols Page. The protocol was updated to include a higher innoculation value from the overnight culture. | We have prepared 42 aliquots of electrocompetent XL1 Blue ''E. coli'' cells using [http://openwetware.org/wiki/IGEM:IMPERIAL/2008/New/Protocols/XL1-Blue_preparation the relevant protocol] listed on our Protocols Page. The protocol was updated to include a higher innoculation value from the overnight culture. | ||
| + | |||
| + | ===Results=== | ||
| + | |||
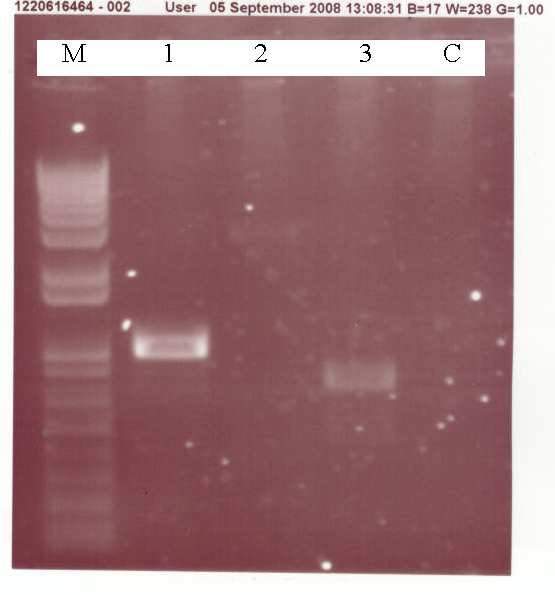
| + | [[Image:4-08PCR.PNG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer. M = Marker, lane 1 is the AmyE 3' Integration sequence, lane 2 the LacI and lane 3 the Aad9 PCR reaction]] | ||
| + | |||
| + | As can be seen, both the AmyE 3' integration sequence and the Aad9 (spectinomycin resistance) gene amplified properly. LacI did not however and so this will need to be repeated. | ||
Revision as of 17:37, 5 September 2008
4 September 2008WetlabCloning
Preparation of Electrocompetent E. coli CellsWe have prepared 42 aliquots of electrocompetent XL1 Blue E. coli cells using the relevant protocol listed on our Protocols Page. The protocol was updated to include a higher innoculation value from the overnight culture. ResultsAs can be seen, both the AmyE 3' integration sequence and the Aad9 (spectinomycin resistance) gene amplified properly. LacI did not however and so this will need to be repeated. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"