Team:TUDelft/Temperature overview
From 2008.igem.org
(→Bacterial Thermometer) |
(→Temperature Overview) |
||
| (31 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
{{Template:TUDelftiGEM2008_menu_home}} | {{Template:TUDelftiGEM2008_menu_home}} | ||
| - | + | =Temperature Overview= | |
| - | = | + | We want to create an RNA-based system that is able to have differential gene expression when temperature changes take place (comparable to the [https://2006.igem.org/Berkeley2006-RiboregulatorsMain Berkeley iGEM 2006] riboregulator lock/key part). |
| + | [[Image:Rna_thermometer.png|200px|right|thumb|'''Figure 1:''' RNA thermometer]] | ||
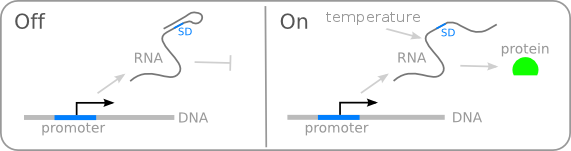
| + | There are several systems suggested in literature that are based on RNA secondary structure <span id="cite_ref_1">[[Team:TUDelft/Temperature_overview#cite_note_1|[1]]]</span>. The idea in general is that if the temperature drops below a certain temperature, the RNA will form stable base-pairs on the Shine-Dalgarno sequence, disabling the ribosome to bind. The base-pairing of this RNA region will block the expression of the protein encoded behind it (figure 1). In this way gene expression can be regulated on the RNA level by temperature. | ||
| - | + | At the beginning of the project, an [[Team:TUDelft/Temperature_analysis|analysis]] has been performed to get more insight into the functioning of an RNA thermometer. The design of the temperature sensitive switch is divided in two phases. In [[Team:TUDelft/Temperature_design|the first phase]] existing RNA thermometers have been turned into BioBrick Standard Biological Parts. In the [[Team:TUDelft/Temperature_design2|second phase]], an RNA thermometer has been redesigned in order to shift the switching temperature using the knowledge gained during the analysis. | |
| - | + | Also some [[Team:TUDelft/Temperature_software|software]] tools have been developed during the project. The first tool, the [[Team:TUDelft/Temperature_software#Stability_Profile_Plotter|Stability Profile Plotter]], has been used during the analysis, to produce plots that characterize the stability of an RNA hairpin structure. The second tool, the [[Team:TUDelft/Temperature_software#RNA_Hairpin_Designer|RNA Hairpin Designer]], provides a (partial) implementation of the design approach as proposed in the second design phase. This tool can be used for the design of temperature sensitive RNA hairpins. | |
| - | + | The results of the labwork indicate that sonication of our samples, obtained the most reliable results. These indicated, as displayed [[Team:TUDelft/Temperature_results#Luciferase_Measurements|here]], that our strain K115035 shows the temperature induced switch-like behavior which was the aim of our project. | |
| - | + | ||
| - | ==== | + | ==References== |
| - | + | ||
| - | = | + | <ol class="references"> |
| - | + | ||
| - | </ | + | <li id="cite_note_1"> [[Team:TUDelft/Temperature_overview#cite_ref_1 | ^]] Narberhaus F. mRNA-mediated detection of environmental conditions. ''Archives of Microbiology'', 178(6):404-410, 2002. [http://www.ncbi.nlm.nih.gov/pubmed/16680139 PMID:16680139]</li> |
| + | |||
| + | </ol> | ||
{{Template:TUDelftiGEM2008_sidebar}} | {{Template:TUDelftiGEM2008_sidebar}} | ||
Latest revision as of 17:54, 29 October 2008
Temperature Overview
We want to create an RNA-based system that is able to have differential gene expression when temperature changes take place (comparable to the Berkeley iGEM 2006 riboregulator lock/key part).
There are several systems suggested in literature that are based on RNA secondary structure [1]. The idea in general is that if the temperature drops below a certain temperature, the RNA will form stable base-pairs on the Shine-Dalgarno sequence, disabling the ribosome to bind. The base-pairing of this RNA region will block the expression of the protein encoded behind it (figure 1). In this way gene expression can be regulated on the RNA level by temperature.
At the beginning of the project, an analysis has been performed to get more insight into the functioning of an RNA thermometer. The design of the temperature sensitive switch is divided in two phases. In the first phase existing RNA thermometers have been turned into BioBrick Standard Biological Parts. In the second phase, an RNA thermometer has been redesigned in order to shift the switching temperature using the knowledge gained during the analysis.
Also some software tools have been developed during the project. The first tool, the Stability Profile Plotter, has been used during the analysis, to produce plots that characterize the stability of an RNA hairpin structure. The second tool, the RNA Hairpin Designer, provides a (partial) implementation of the design approach as proposed in the second design phase. This tool can be used for the design of temperature sensitive RNA hairpins.
The results of the labwork indicate that sonication of our samples, obtained the most reliable results. These indicated, as displayed here, that our strain K115035 shows the temperature induced switch-like behavior which was the aim of our project.
References
- ^ Narberhaus F. mRNA-mediated detection of environmental conditions. Archives of Microbiology, 178(6):404-410, 2002. [http://www.ncbi.nlm.nih.gov/pubmed/16680139 PMID:16680139]
 "
"