Team:TUDelft/Temperature results
From 2008.igem.org
(Difference between revisions)
(→Results) |
(→Results) |
||
| Line 7: | Line 7: | ||
The first challenge of this project was to assemble all the devices we wanted to measure. An overview of these devices can be found [https://2008.igem.org/Team:TUDelft/Temperature_testing here]. For the actual construction of all devices the 3A assembly strategy was chosen. A complete protocol of this strategy can be found on [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly OpenWetWare]. A schematic representation of this assembly method is depicted in figure 1A and 1B.<br/><br/> | The first challenge of this project was to assemble all the devices we wanted to measure. An overview of these devices can be found [https://2008.igem.org/Team:TUDelft/Temperature_testing here]. For the actual construction of all devices the 3A assembly strategy was chosen. A complete protocol of this strategy can be found on [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly OpenWetWare]. A schematic representation of this assembly method is depicted in figure 1A and 1B.<br/><br/> | ||
| - | [[Image:TUDelft3Amixture.png|thumb|250px|left|Figure 1A. The mixture present in the ligation mix before it is ligated during 3A assembly. The black box indicates the [http://partsregistry.org/Part:BBa_P1010 CcdB 'death' gene]. The arrows in red, blue and green indicate different types of antibiotic resistance. Restriction sites are indicated by: E = ''Eco''R1; P = ''Pst''I; X = ''Xba''I; S = ''Spe''I]] | + | [[Image:TUDelft3Amixture.png|thumb|250px|left|Figure 1A. The mixture present in the ligation mix before it is ligated during 3A assembly. The black box indicates the [http://partsregistry.org/Part:BBa_P1010 CcdB 'death' gene]. The arrows in red, blue and green indicate different types of antibiotic resistance. Restriction sites are indicated by: E = ''Eco''R1; P = ''Pst''I; X = ''Xba''I; S = ''Spe''I. Picture taken from OpenWetWare, http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly OpenWetWare]] |
| - | [[Image:TUDelftschematic3A.png|thumb|250px|Figure 1B. Representation of the 3A assembly method. The black arrow indicates the CcdB gene. The arrows in other colors indicate different types of antibiotic resistance. Restriction sites are indicated by: E = ''Eco''R1; P = ''Pst''I; X = ''Xba''I; S = ''Spe''I]] | + | [[Image:TUDelftschematic3A.png|thumb|250px|Figure 1B. Representation of the 3A assembly method. The black arrow indicates the CcdB gene. The arrows in other colors indicate different types of antibiotic resistance. Restriction sites are indicated by: E = ''Eco''R1; P = ''Pst''I; X = ''Xba''I; S = ''Spe''I. Picture taken from OpenWetWare, http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly OpenWetWare]] |
{{clear}} | {{clear}} | ||
Revision as of 11:36, 28 October 2008
Results
The first challenge of this project was to assemble all the devices we wanted to measure. An overview of these devices can be found here. For the actual construction of all devices the 3A assembly strategy was chosen. A complete protocol of this strategy can be found on OpenWetWare. A schematic representation of this assembly method is depicted in figure 1A and 1B.
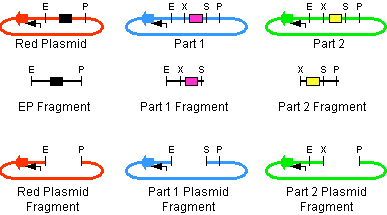
Figure 1A. The mixture present in the ligation mix before it is ligated during 3A assembly. The black box indicates the CcdB 'death' gene. The arrows in red, blue and green indicate different types of antibiotic resistance. Restriction sites are indicated by: E = EcoR1; P = PstI; X = XbaI; S = SpeI. Picture taken from OpenWetWare, http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly OpenWetWare
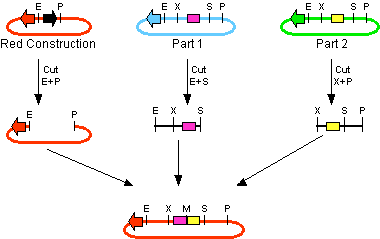
Figure 1B. Representation of the 3A assembly method. The black arrow indicates the CcdB gene. The arrows in other colors indicate different types of antibiotic resistance. Restriction sites are indicated by: E = EcoR1; P = PstI; X = XbaI; S = SpeI. Picture taken from OpenWetWare, http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly OpenWetWare
For the succesful use of this assembly, it is essential that the DB3.1 Eschirichia coli strain is not used as it tolerates the presence of the CcdB gene. Throughout the project we used commercial Top10 cells of Invitrogen that were made competent chemically. Furthermore, it is important to note that the XbaI and SpeI restriction enzymes generate compatible sticky ends.
 "
"